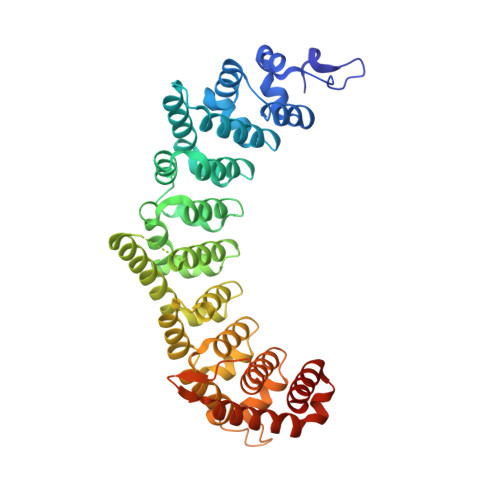
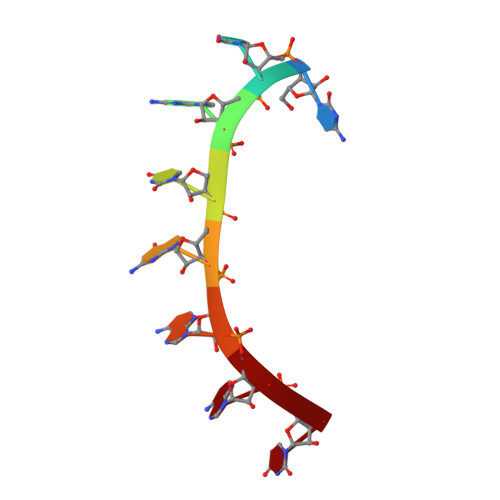
A crystal structure of a collaborative RNA regulatory complex reveals mechanisms to refine target specificity.
Qiu, C., Bhat, V.D., Rajeev, S., Zhang, C., Lasley, A.E., Wine, R.N., Campbell, Z.T., Tanaka Hall, T.M.T.(2019) Elife 8
- PubMed: 31397673
- DOI: https://doi.org/10.7554/eLife.48968
- Primary Citation of Related Structures:
6PUN - PubMed Abstract:
In the Caenorhabditis elegans germline, fem-3 Binding Factor (FBF) partners with LST-1 to maintain stem cells. A crystal structure of an FBF-2/LST-1/RNA complex revealed that FBF-2 recognizes a short RNA motif different from the characteristic 9-nt FBF binding element, and compact motif recognition coincided with curvature changes in the FBF-2 scaffold. Previously, we engineered FBF-2 to favor recognition of shorter RNA motifs without curvature change (Bhat et al., 2019). In vitro selection of RNAs bound by FBF-2 suggested sequence specificity in the central region of the compact element. This bias, reflected in the crystal structure, was validated in RNA-binding assays. FBF-2 has the intrinsic ability to bind to this shorter motif. LST-1 weakens FBF-2 binding affinity for short and long motifs, which may increase target selectivity. Our findings highlight the role of FBF scaffold flexibility in RNA recognition and suggest a new mechanism by which protein partners refine target site selection.
Organizational Affiliation:
Epigenetics and Stem Cell Biology Laboratory, National Institute of Environmental Health Sciences, National Institutes of Health, Research Triangle Park, United States.