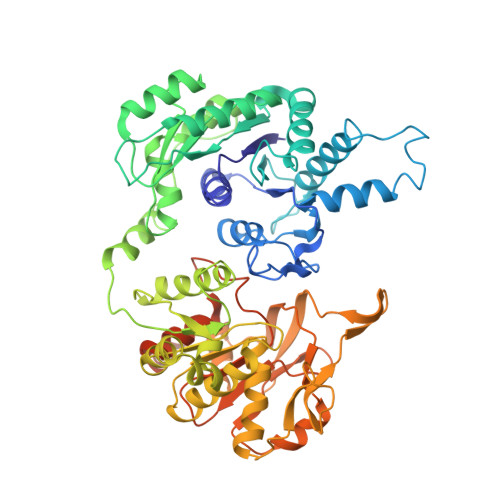
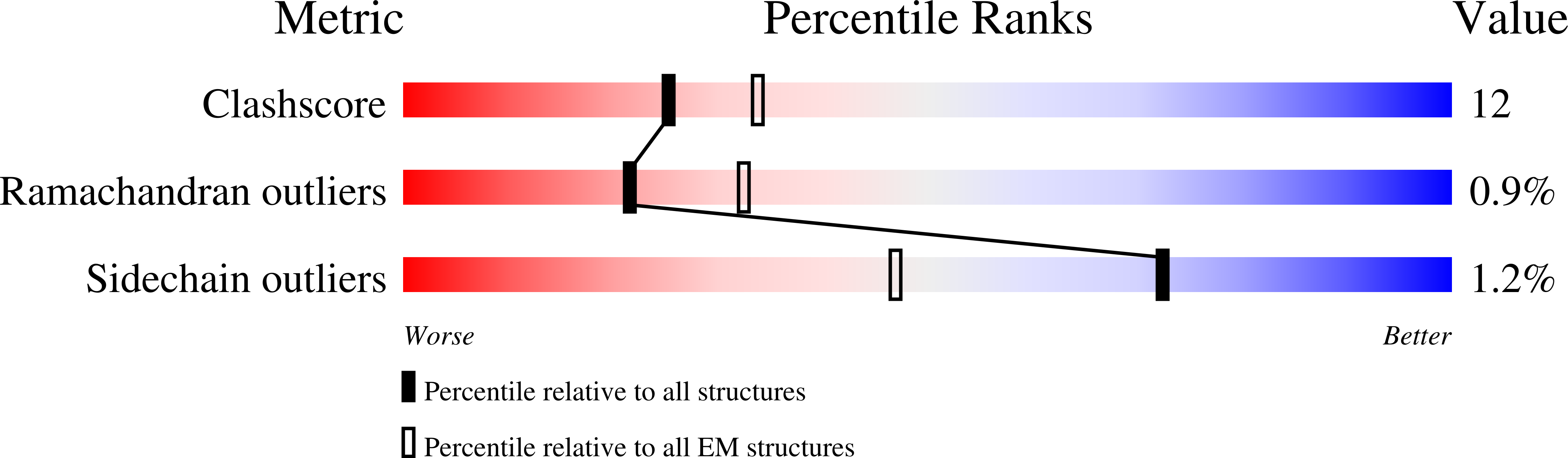Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments.
Lynch, E.M., Kollman, J.M.(2020) Nat Struct Mol Biol 27: 42-48
- PubMed: 31873303
- DOI: https://doi.org/10.1038/s41594-019-0352-5
- Primary Citation of Related Structures:
6PK4, 6PK7 - PubMed Abstract:
Many enzymes assemble into defined oligomers, providing a mechanism for cooperatively regulating activity. Recent studies have described a mode of regulation in which enzyme activity is modulated by polymerization into large-scale filaments. Here we describe an ultrasensitive form of polymerization-based regulation employed by human CTP synthase 2 (CTPS2). Cryo-EM structures reveal that CTPS2 filaments dynamically switch between active and inactive forms in response to changes in substrate and product levels. Linking the conformational state of many CTPS2 subunits in a filament results in highly cooperative regulation, greatly exceeding the limits of cooperativity for the CTPS2 tetramer alone. The structures reveal a link between conformation and control of ammonia channeling between the enzyme's active sites, and explain differences in regulation of human CTPS isoforms. This filament-based mechanism of enhanced cooperativity demonstrates how the widespread phenomenon of enzyme polymerization can be adapted to achieve different regulatory outcomes.
Organizational Affiliation:
Department of Biochemistry, University of Washington, Seattle, WA, USA.