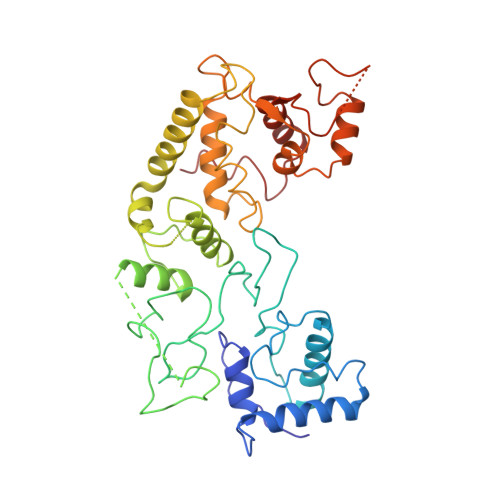
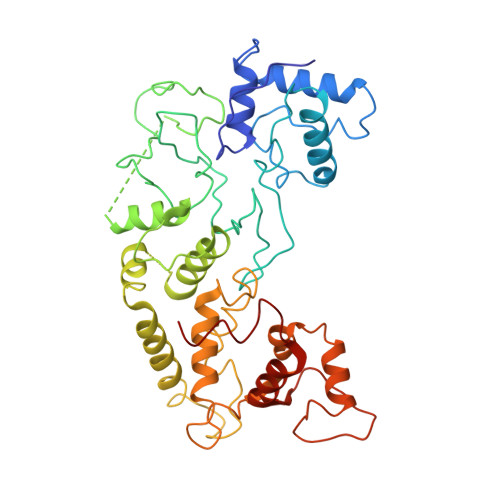
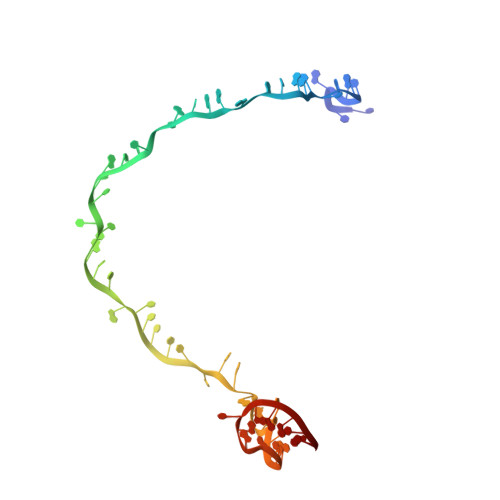
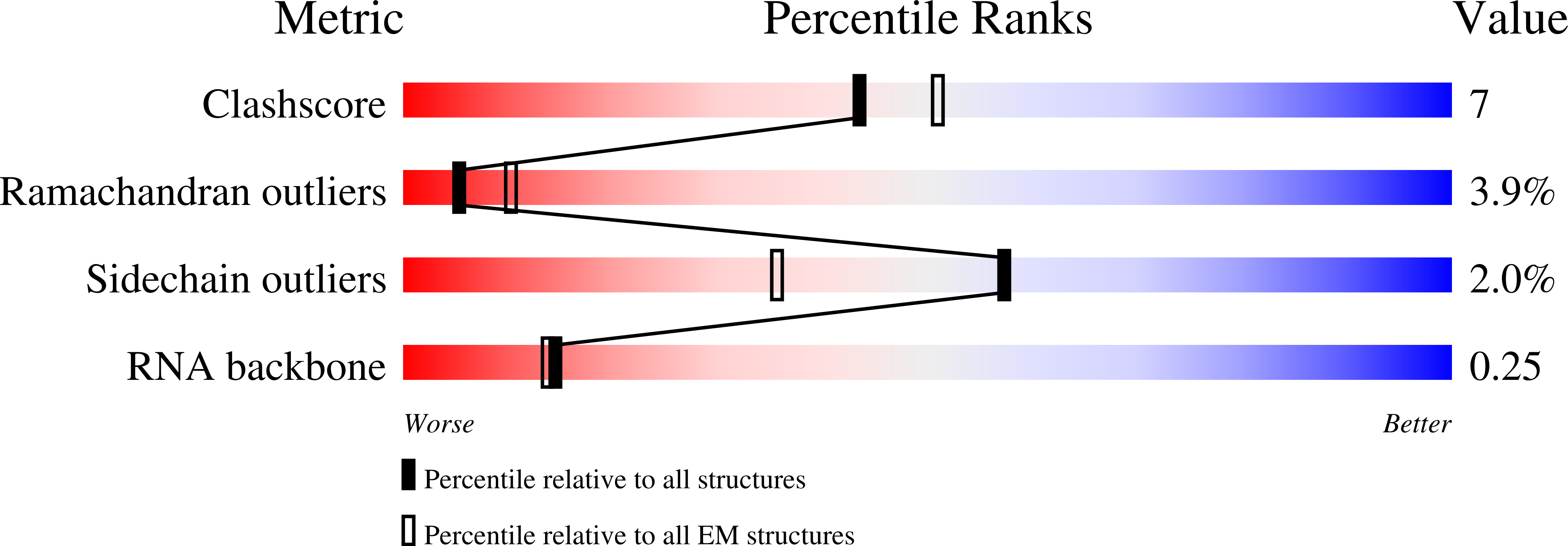Structural basis of DNA targeting by a transposon-encoded CRISPR-Cas system.
Halpin-Healy, T.S., Klompe, S.E., Sternberg, S.H., Fernandez, I.S.(2020) Nature 577: 271-274
- PubMed: 31853065
- DOI: https://doi.org/10.1038/s41586-019-1849-0
- Primary Citation of Related Structures:
6PIF, 6PIG, 6PIJ - PubMed Abstract:
Bacteria use adaptive immune systems encoded by CRISPR and Cas genes to maintain genomic integrity when challenged by pathogens and mobile genetic elements 1-3 . Type I CRISPR-Cas systems typically target foreign DNA for degradation via joint action of the ribonucleoprotein complex Cascade and the helicase-nuclease Cas3 4,5 , but nuclease-deficient type I systems lacking Cas3 have been repurposed for RNA-guided transposition by bacterial Tn7-like transposons 6,7 . How CRISPR- and transposon-associated machineries collaborate during DNA targeting and insertion remains unknown. Here we describe structures of a TniQ-Cascade complex encoded by the Vibrio cholerae Tn6677 transposon using cryo-electron microscopy, revealing the mechanistic basis of this functional coupling. The cryo-electron microscopy maps enabled de novo modelling and refinement of the transposition protein TniQ, which binds to the Cascade complex as a dimer in a head-to-tail configuration, at the interface formed by Cas6 and Cas7 near the 3' end of the CRISPR RNA (crRNA). The natural Cas8-Cas5 fusion protein binds the 5' crRNA handle and contacts the TniQ dimer via a flexible insertion domain. A target DNA-bound structure reveals critical interactions necessary for protospacer-adjacent motif recognition and R-loop formation. This work lays the foundation for a structural understanding of how DNA targeting by TniQ-Cascade leads to downstream recruitment of additional transposase proteins, and will guide protein engineering efforts to leverage this system for programmable DNA insertions in genome-engineering applications.
Organizational Affiliation:
Department of Biochemistry and Molecular Biophysics, Columbia University, New York, NY, USA.