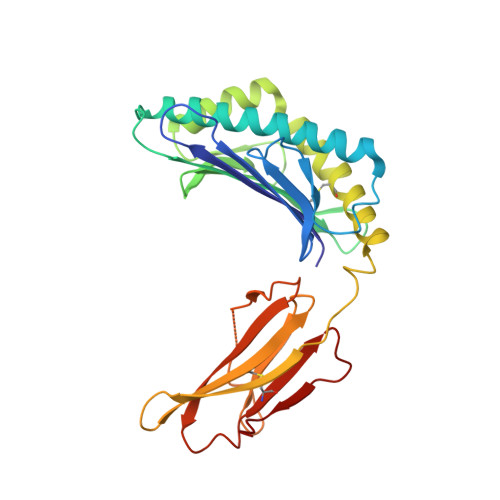
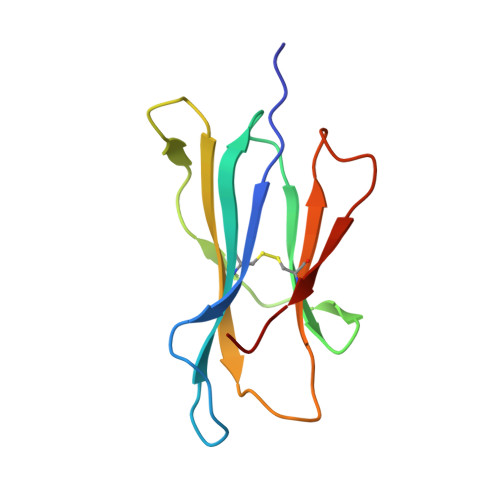
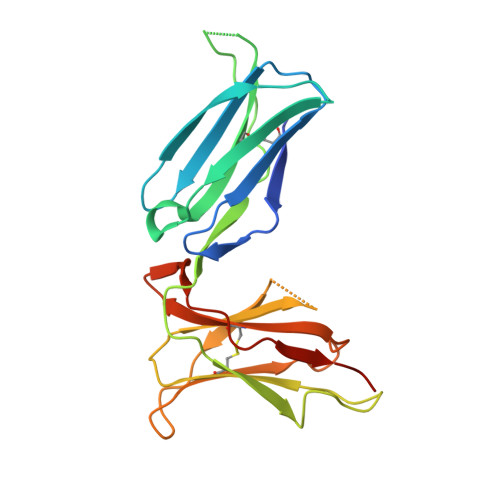
Structural plasticity of KIR2DL2 and KIR2DL3 enables altered docking geometries atop HLA-C.
Moradi, S., Stankovic, S., O'Connor, G.M., Pymm, P., MacLachlan, B.J., Faoro, C., Retiere, C., Sullivan, L.C., Saunders, P.M., Widjaja, J., Cox-Livingstone, S., Rossjohn, J., Brooks, A.G., Vivian, J.P.(2021) Nat Commun 12: 2173-2173
- PubMed: 33846289
- DOI: https://doi.org/10.1038/s41467-021-22359-x
- Primary Citation of Related Structures:
6PA1, 6PAG - PubMed Abstract:
The closely related inhibitory killer-cell immunoglobulin-like receptors (KIR), KIR2DL2 and KIR2DL3, regulate the activation of natural killer cells (NK) by interacting with the human leukocyte antigen-C1 (HLA-C1) group of molecules. KIR2DL2, KIR2DL3 and HLA-C1 are highly polymorphic, with this variation being associated with differences in the onset and progression of some human diseases. However, the molecular bases underlying these associations remain unresolved. Here, we determined the crystal structures of KIR2DL2 and KIR2DL3 in complex with HLA-C*07:02 presenting a self-epitope. KIR2DL2 differed from KIR2DL3 in docking modality over HLA-C*07:02 that correlates with variabilty of recognition of HLA-C1 allotypes. Mutagenesis assays indicated differences in the mechanism of HLA-C1 allotype recognition by KIR2DL2 and KIR2DL3. Similarly, HLA-C1 allotypes differed markedly in their capacity to inhibit activation of primary NK cells. These functional differences derive, in part, from KIR2DS2 suggesting KIR2DL2 and KIR2DL3 binding geometries combine with other factors to distinguish HLA-C1 functional recognition.
Organizational Affiliation:
Infection and Immunity Program and Department of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC, Australia.