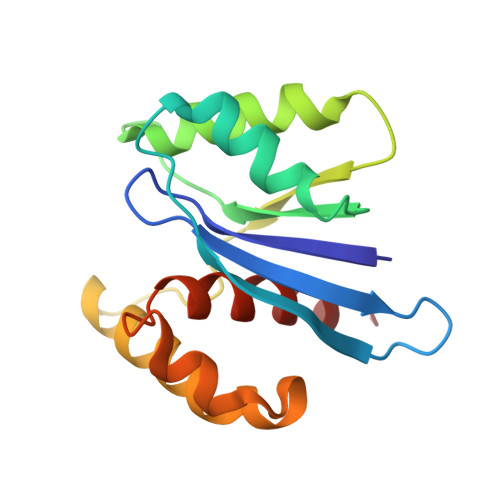
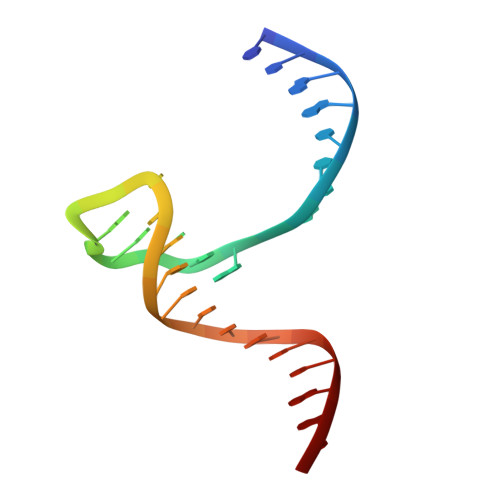
Structural insights into the promiscuous DNA binding and broad substrate selectivity of fowlpox virus resolvase.
Li, N., Shi, K., Rao, T., Banerjee, S., Aihara, H.(2020) Sci Rep 10: 393-393
- PubMed: 31941902
- DOI: https://doi.org/10.1038/s41598-019-56825-w
- Primary Citation of Related Structures:
6P7A, 6P7B - PubMed Abstract:
Fowlpox virus resolvase (Fpr) is an endonuclease that cleaves a broad range of branched DNA structures, including the Holliday junction (HJ), with little sequence-specificity. To better understand the mechanisms underlying its relaxed substrate specificity, we determined the crystal structures of Fpr and that in a novel complex with HJ at 3.1-Å resolution. In the Fpr-HJ complex, two Fpr dimers use several distinct regions to interact with different DNA structural motifs, showing versatility in DNA-binding. Biochemical and solution NMR data support the existence of non-canonical modes of HJ interaction in solution. The binding of Fpr to various DNA motifs are mediated by its flat DNA-binding surface, which is centered on a short loop spanning K61 to I72 and flanked by longer α-helices at the outer edges, and basic side grooves near the dimer interface. Replacing the Fpr loop K61~I72 with a longer loop from Thermus thermophilus RuvC (E71~A87) endows Fpr with an enhanced selectivity toward HJ cleavage but with a target sequence preference distinct from that of RuvC, highlighting a unique role of this loop region in Fpr-HJ interaction. Our work helps explain the broad substrate selectivity of Fpr and suggests a possible mode of its association with poxvirus hairpin telomeres.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology, and Biophysics, University of Minnesota, 6-155 Jackson Hall, 321 Church Street S.E. Minneapolis, Minneapolis, MN, 55455, USA.