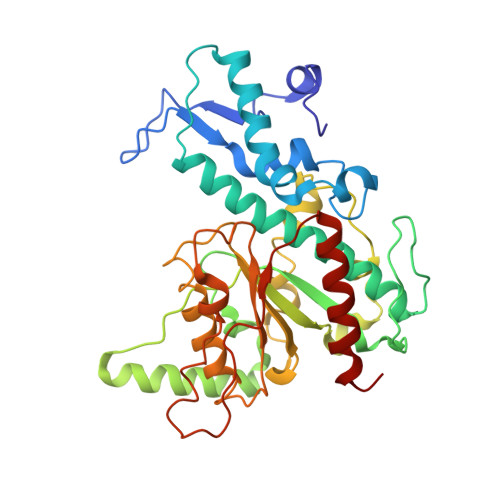
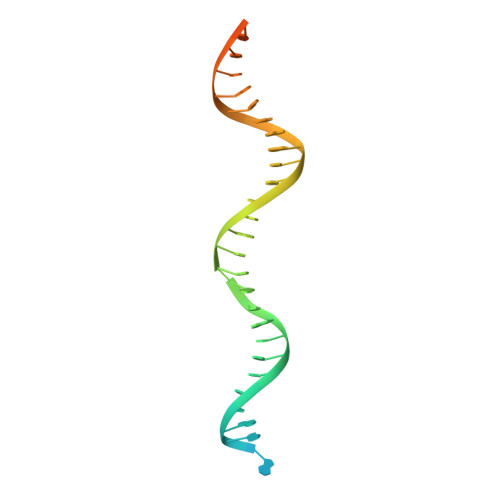
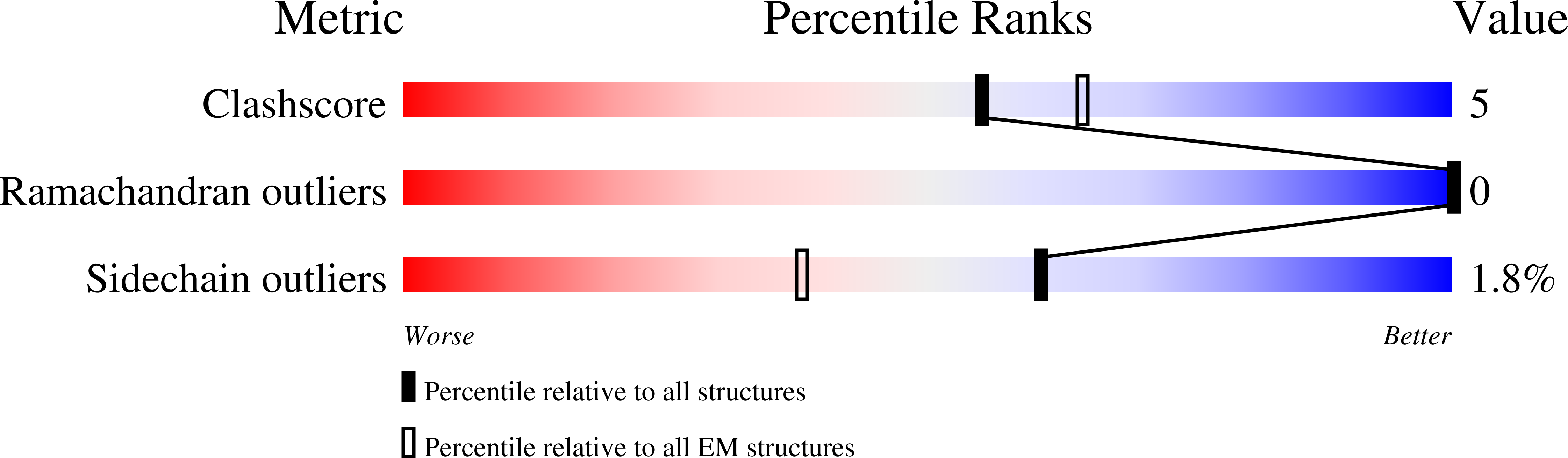Mechanism of Filamentation-Induced Allosteric Activation of the SgrAI Endonuclease.
Polley, S., Lyumkis, D., Horton, N.C.(2019) Structure 27: 1497-1507.e3
- PubMed: 31447289
- DOI: https://doi.org/10.1016/j.str.2019.08.001
- Primary Citation of Related Structures:
6OBJ - PubMed Abstract:
Filament formation by enzymes is increasingly recognized as an important phenomenon with potentially unique regulatory properties and biological roles. SgrAI is an allosterically regulated type II restriction endonuclease that forms filaments with enhanced DNA cleavage activity and altered sequence specificity. Here, we present the cryoelectron microscopy (cryo-EM) structure of the filament of SgrAI in its activated configuration. The structural data illuminate the mechanistic origin of hyperaccelerated DNA cleavage activity and suggests how indirect DNA sequence readout within filamentous SgrAI may enable recognition of substantially more nucleotide sequences than its low-activity form, thereby altering and partially relaxing its DNA sequence specificity. Together, substrate DNA binding, indirect readout, and filamentation simultaneously enhance SgrAI's catalytic activity and modulate substrate preference. This unusual enzyme mechanism may have evolved to perform the specialized functions of bacterial innate immunity in rapid defense against invading phage DNA without causing damage to the host DNA.
Organizational Affiliation:
Department of Biophysics, Bose Institute, Kolkata 700054, India.