Structural determinants for accurate dephosphorylation of RNA polymerase II by its cognate C-terminal domain (CTD) phosphatase during eukaryotic transcription.
Irani, S., Sipe, S.N., Yang, W., Burkholder, N.T., Lin, B., Sim, K., Matthews, W.L., Brodbelt, J.S., Zhang, Y.(2019) J Biol Chem 294: 8592-8605
- PubMed: 30971428
- DOI: https://doi.org/10.1074/jbc.RA119.007697
- Primary Citation of Related Structures:
6NPW - PubMed Abstract:
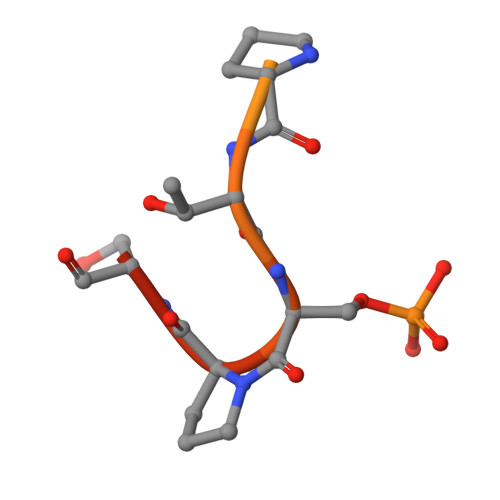
The C-terminal domain (CTD) of RNA polymerase II contains a repetitive heptad sequence (YSPTSPS) whose phosphorylation states coordinate eukaryotic transcription by recruiting protein regulators. The precise placement and removal of phosphate groups on specific residues of the CTD are critical for the fidelity and effectiveness of RNA polymerase II-mediated transcription. During transcriptional elongation, phosphoryl-Ser 5 (pSer 5 ) is gradually dephosphorylated by CTD phosphatases, whereas Ser 2 phosphorylation accumulates. Using MS, X-ray crystallography, protein engineering, and immunoblotting analyses, here we investigated the structure and function of SSU72 homolog, RNA polymerase II CTD phosphatase (Ssu72, from Drosophila melanogaster ), an essential CTD phosphatase that dephosphorylates pSer 5 at the transition from elongation to termination, to determine the mechanism by which Ssu72 distinguishes the highly similar pSer 2 and pSer 5 CTDs. We found that Ssu72 dephosphorylates pSer 5 effectively but only has low activities toward pSer 7 and pSer 2 The structural analysis revealed that Ssu72 requires that the proline residue in the substrate's SP motif is in the cis configuration, forming a tight β-turn for recognition by Ssu72. We also noted that residues flanking the SP motif, such as the bulky Tyr 1 next to Ser 2 , prevent the formation of such configuration and enable Ssu72 to distinguish among the different SP motifs. The phosphorylation of Tyr 1 further prohibited Ssu72 binding to pSer 2 and thereby prevented untimely Ser 2 dephosphorylation. Our results reveal critical roles for Tyr 1 in differentiating the phosphorylation states of Ser 2 /Ser 5 of CTD in RNA polymerase II that occur at different stages of transcription.
Organizational Affiliation:
Department of Chemical Engineering, The University of Texas, Austin, Texas 78712; Department of Molecular Biosciences, The University of Texas, Austin, Texas 78712.