Cisplatin protects mice from challenge ofCryptococcus neoformansby targeting the Prp8 intein.
Li, Z., Fu, B., Green, C.M., Liu, B., Zhang, J., Lang, Y., Chaturvedi, S., Belfort, M., Liao, G., Li, H.(2019) Emerg Microbes Infect 8: 895-908
- PubMed: 31223062
- DOI: https://doi.org/10.1080/22221751.2019.1625727
- Primary Citation of Related Structures:
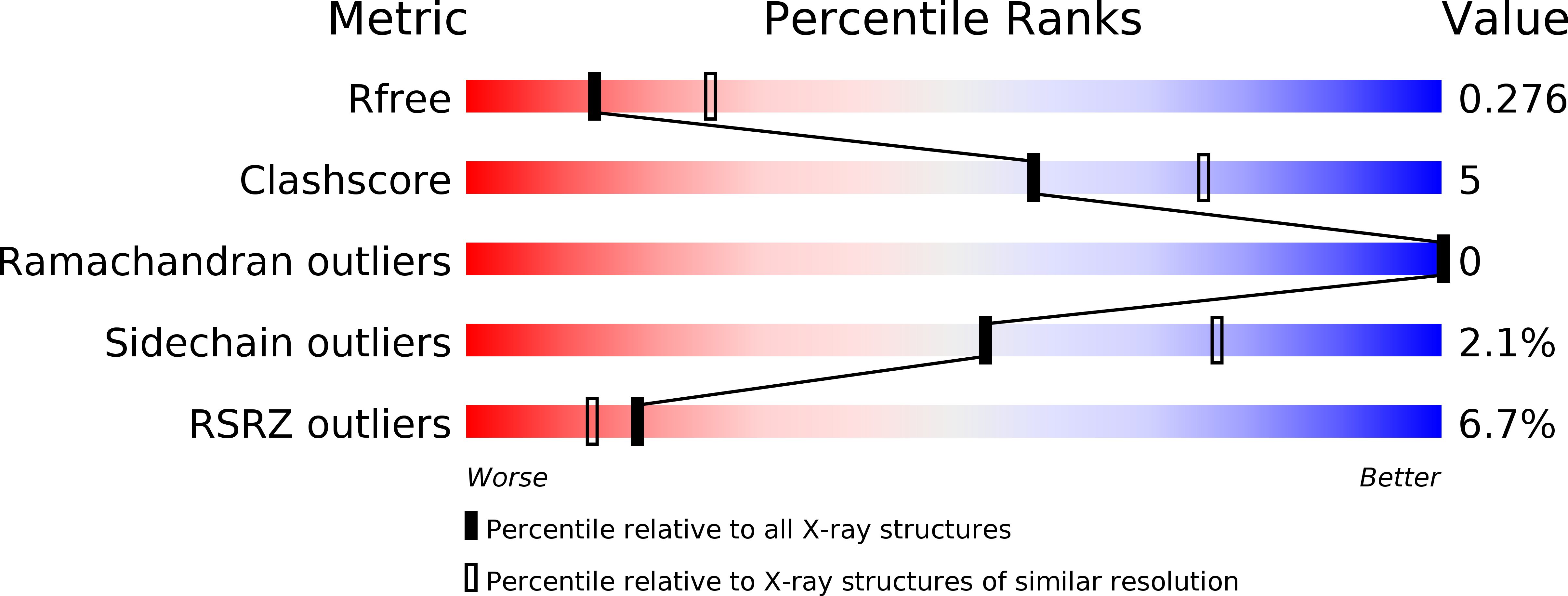
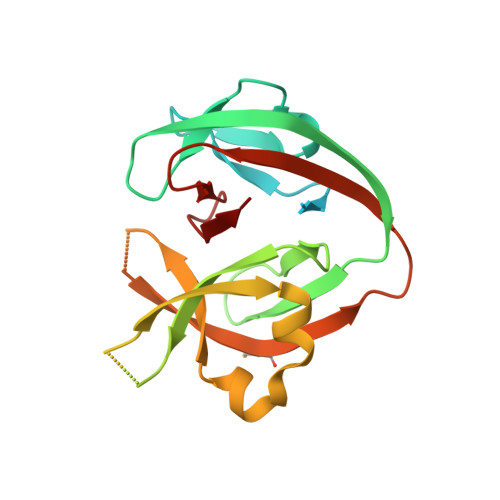
6MWY, 6MYL - PubMed Abstract:
The Prp8 intein is one of the most widespread eukaryotic inteins, present in important pathogenic fungi, including Cryptococcus and Aspergillus species. Because the processed Prp8 carries out essential and non-redundant cellular functions, a Prp8 intein inhibitor is a mechanistically novel antifungal agent. In this report, we demonstrated that cisplatin, an FDA-approved cancer drug, significantly arrested growth of Prp8 intein-containing fungi C. neoformans and C. gattii, but only poorly inhibited growth of intein-free Candida species. These results suggest that cisplatin arrests fungal growth through specific inhibition of the Prp8 intein. Cisplatin was also found to significantly inhibit growth of C. neoformans in a mouse model. Our results further showed that cisplatin inhibited Prp8 intein splicing in vitro in a dose-dependent manner by direct binding to the Prp8 intein. Crystal structures of the apo- and cisplatin-bound Prp8 inteins revealed that two degenerate cisplatin molecules bind at the intein active site. Mutation of the splicing-site residues led to loss of cisplatin binding, as well as impairment of intein splicing. Finally, we found that overexpression of the Prp8 intein in cryptococcal species conferred cisplatin resistance. Overall, these results indicate that the Prp8 intein is a novel antifungal target worth further investigation.
Organizational Affiliation:
New York State Department of Health , Wadsworth Center, Albany, NY, USA.