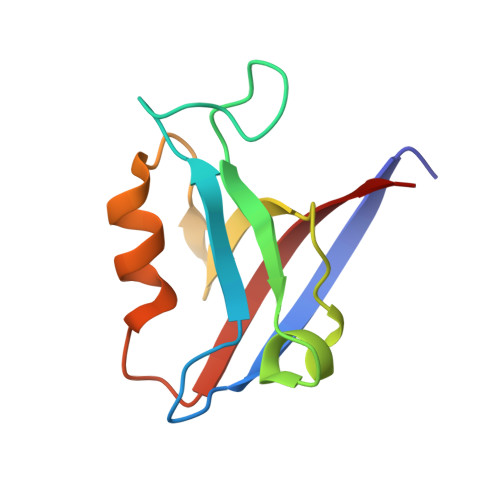
Crystal structure of the human Scribble PDZ1 domain bound to the PDZ-binding motif of APC.
How, J.Y., Caria, S., Humbert, P.O., Kvansakul, M.(2019) FEBS Lett 593: 533-542
- PubMed: 30659601
- DOI: https://doi.org/10.1002/1873-3468.13329
- Primary Citation of Related Structures:
6MS1 - PubMed Abstract:
Scribble (SCRIB) is an important adaptor protein that controls the establishment and maintenance of apico-basal cell polarity. To better understand how SCRIB controls cell polarity signalling via its PDZ domains, we investigated human SCRIB interactions with adenomatous polyposis coli (APC). We show that SCRIB PDZ1, PDZ2 and PDZ3 are the major interactors with the APC PDZ-binding motif (PBM), whereas SCRIB PDZ4 does not show detectable binding to APC. We then determined the crystal structure of SCRIB PDZ1 domain bound to the APC PBM. Our findings reveal a previously unreported pattern of interactions between the SCRIB PDZ domain region with the C-terminal PDZ binding motif of APC, where SCRIB PDZ1 domain is the highest affinity site.
Organizational Affiliation:
Department of Biochemistry & Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Australia.