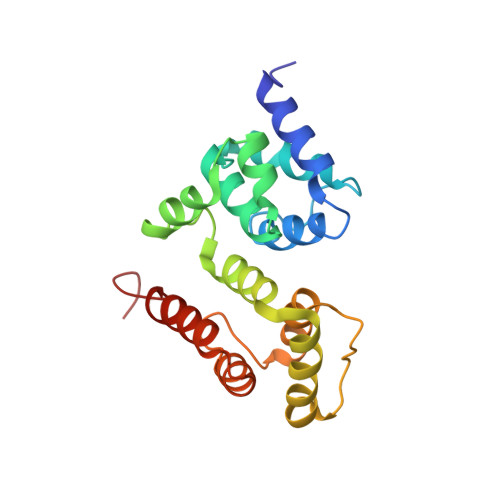
Solution structure of c-FLIP death effector domains.
Bai, Z.Q., Ma, X., Liu, B., Huang, T., Hu, K.(2022) Biochem Biophys Res Commun 617: 1-6
- PubMed: 35688044
- DOI: https://doi.org/10.1016/j.bbrc.2022.05.086
- Primary Citation of Related Structures:
6M6O - PubMed Abstract:
The formation of death-inducing signaling complex (DISC) and death effector domain (DED) filament initiates extrinsic apoptosis. Recruitment and activation of procaspase-8 at the DISC are regulated by c-FLIP. The interaction between c-FLIP and procaspase-8 is mediated by their tandem DEDs (tDED). However, the structure of c-FLIP tDED and how c-FLIP interferes with procaspase-8 activation at the DISC remain elusive. Here, we solved the monomeric structure of c-FLIP tDED (F114G) at near physiological pH by solution nuclear magnetic resonance (NMR). Structural superimposition reveals c-FLIP tDED (F114G) adopts a structural topology similar to that of procaspase-8 tDED . Our results provide a structural basis for understanding how c-FLIP interacts with procaspase-8 and the molecular mechanisms of c-FLIP in regulating cell death.
Organizational Affiliation:
State Key Laboratory of Phytochemistry and Plant Resources in West China, Kunming Institute of Botany, Chinese Academy of Sciences, 132 Lanhei Road, Heilongtan, Kunming, 650201, Yunnan, China; Innovative Institute of Chinese Medicine and Pharmacy, Chengdu University of Traditional Chinese Medicine, Chengdu, 611137, China; University of Chinese Academy of Sciences, Beijing, 100049, China.