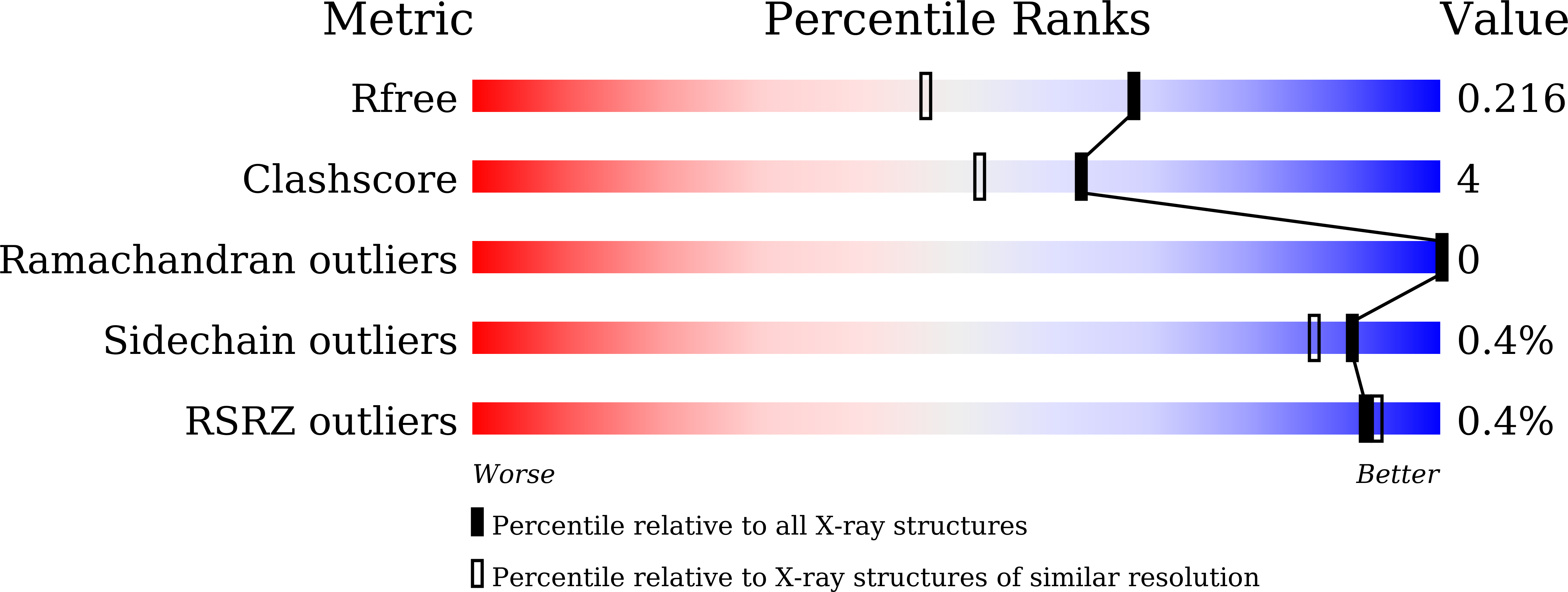
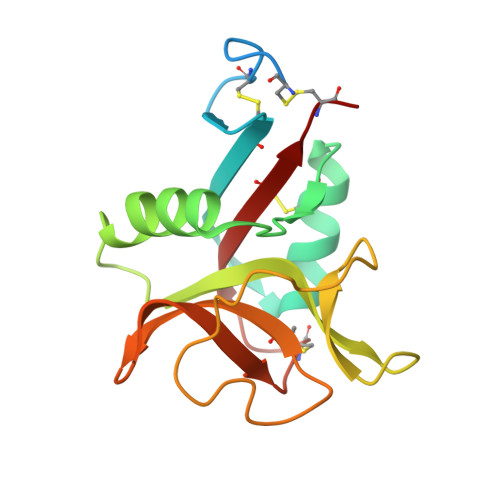
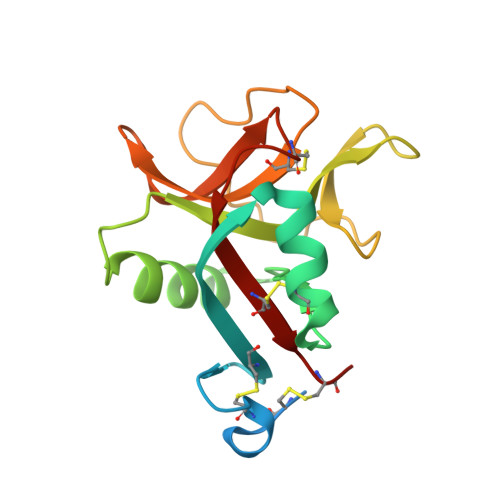
Novel carbohydrate-recognition mode of the invertebrate C-type lectin SPL-1 from Saxidomus purpuratus revealed by the GlcNAc-complex crystal in the presence of Ca2.
Unno, H., Higuchi, S., Goda, S., Hatakeyama, T.(2020) Acta Crystallogr F Struct Biol Commun 76: 271-277
- PubMed: 32510468
- DOI: https://doi.org/10.1107/S2053230X20007256
- Primary Citation of Related Structures:
6M5M - PubMed Abstract:
The C-type lectins SPL-1 and SPL-2 from the bivalve Saxidomus purpuratus are composed of A and B chains and of two B chains, respectively. They bind specific carbohydrates containing acetamido groups, such as N-acetylglucosamine (GlcNAc) and N-acetylgalactosamine (GalNAc), in a Ca 2+ -independent manner. Unlike ordinary C-type lectins, which require Ca 2+ ions for carbohydrate recognition, these lectins recognize specific carbohydrates mainly through interactions with the acetamido group without Ca 2+ ions, even though Ca 2+ enhances the binding affinity of these lectins, especially SPL-1. In the present study, the crystal structure of the SPL-1-GlcNAc complex in the presence of Ca 2+ revealed that the binding of SPL-1 to GlcNAc is stabilized by hydrogen bonds to the water molecule(s) coordinating Ca 2+ , whereas in ordinary C-type lectins Ca 2+ directly forms coordinate bonds to the hydroxy groups of carbohydrates. These differences may also allow SPL-1 and SPL-2 to recognize both GlcNAc and GalNAc, which have different orientations of the 4-hydroxy group.
Organizational Affiliation:
Biomolecular Chemistry Laboratory, Graduate School of Engineering, Nagasaki University, 1-14 Bunkyo-machi, Nagasaki 852-8521, Japan.