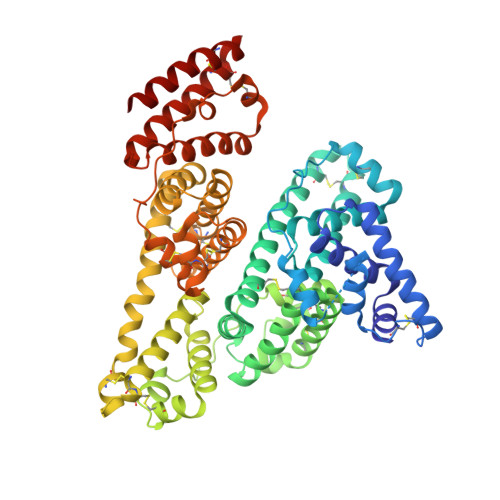
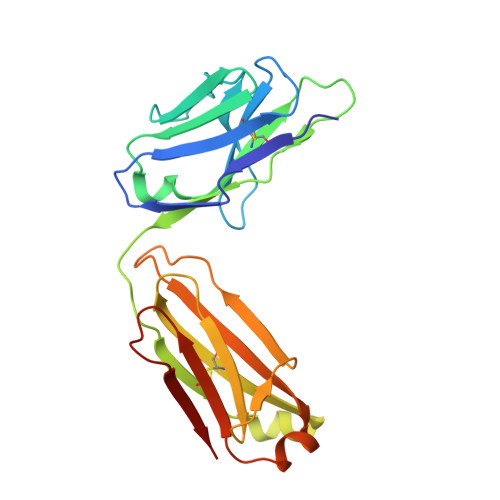
Structural basis of serum albumin recognition by SL335, an antibody Fab extending the serum half-life of protein therapeutics.
Cho, S.Y., Han, J., Cha, S.H., Yoon, S.I.(2020) Biochem Biophys Res Commun 526: 941-946
- PubMed: 32284170
- DOI: https://doi.org/10.1016/j.bbrc.2020.03.133
- Primary Citation of Related Structures:
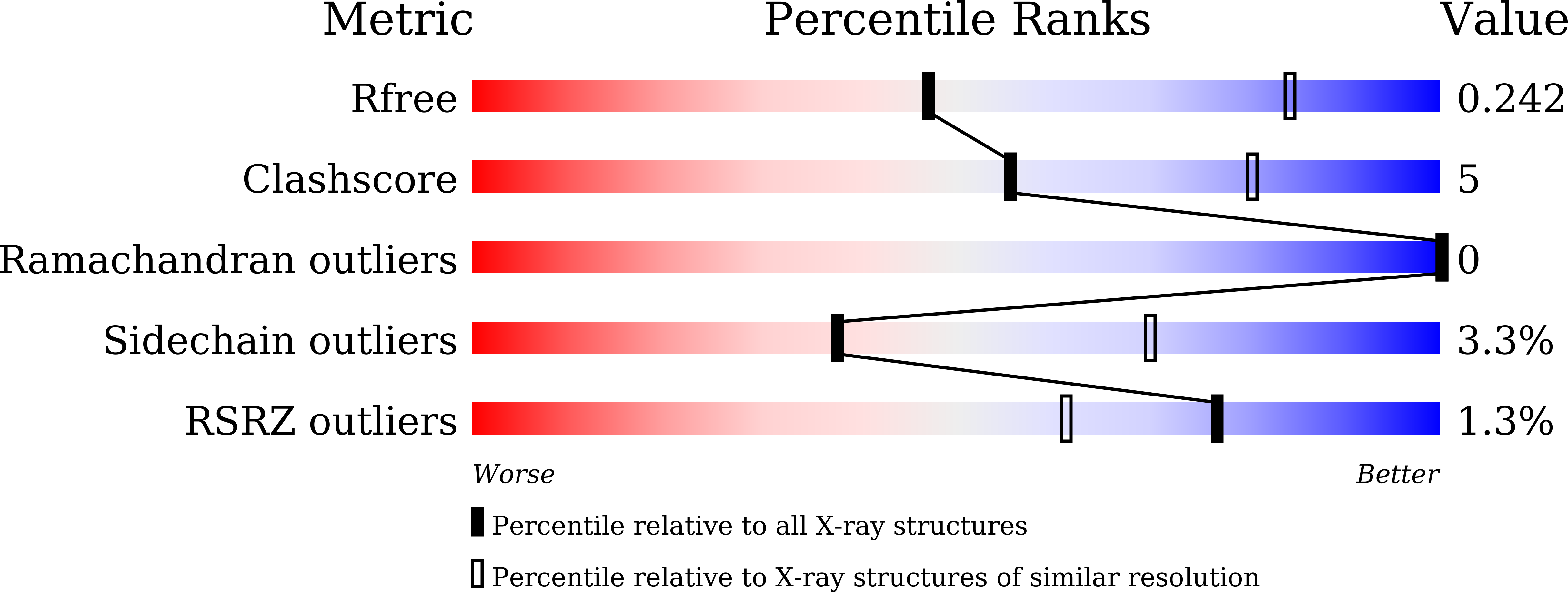
6M58 - PubMed Abstract:
Human serum albumin (HSA) has been used to extend the serum half-lives of various protein therapeutics through genetic fusion because HSA exhibits an exceptionally long circulation time as a result of neonatal Fc receptor (FcRn)-mediated recycling. As another serum half-life extender, the human antibody Fab SL335 that strongly binds HSA was developed. When SL335 was fused to a protein therapeutic, SL335 was shown to prolong the half-life of the drug. Despite the significance of SL335-HSA binding in the extension of drug circulation time, it remains unclear how SL335 interacts with HSA at a molecular structural level. To reveal the structural basis of HSA recognition by SL335, we determined the crystal structure of the SL335-HSA complex at a resolution of 2.95 Å. SL335 binds HSA at a 1:1 stoichiometry. SL335 uses the exposed loops of its heavy and light chains to specifically recognize the IIa and IIb subdomains of HSA. The SL335 epitope is located on the opposite side of the FcRn-binding site and does not overlap with it, suggesting that SL335 extends the serum half-lives of itself and its fusion partner through an FcRn-dependent recycling mechanism.
Organizational Affiliation:
Division of Biomedical Convergence, College of Biomedical Science, Kangwon National University, Chuncheon, 24341, Republic of Korea.