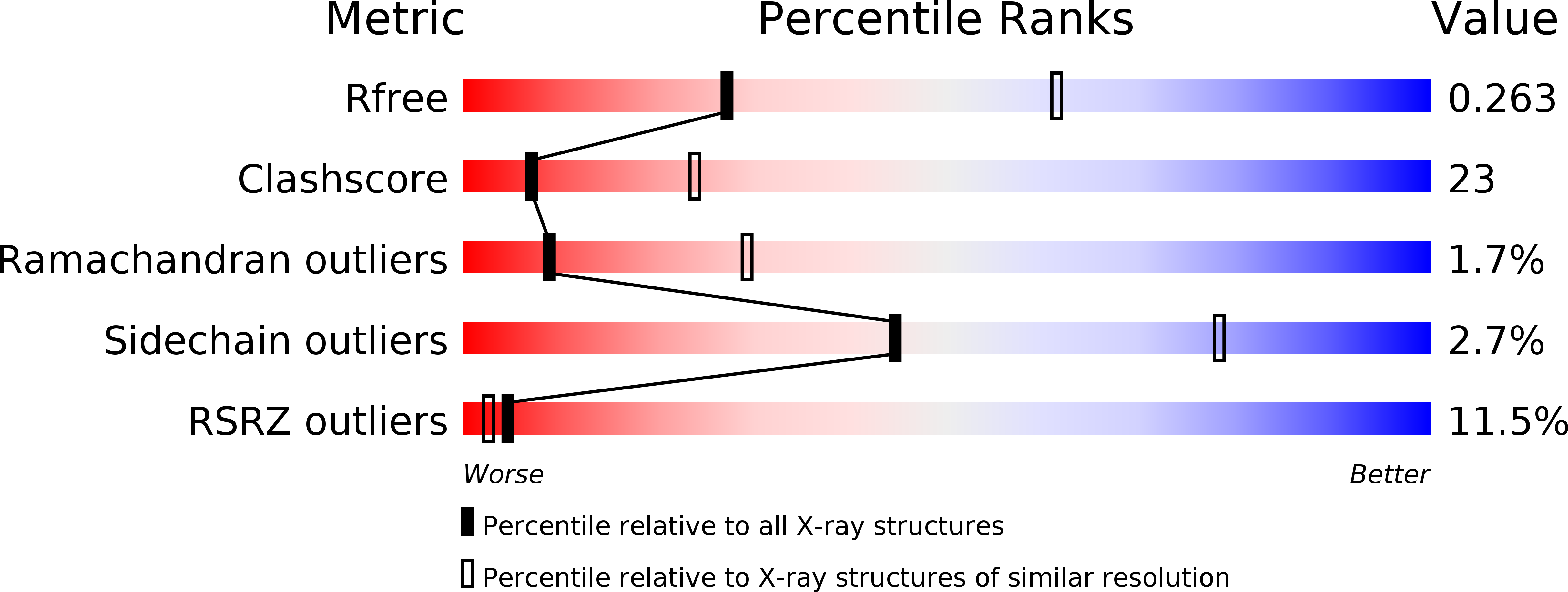
Mechanistic insights into m6A modification of U6 snRNA by human METTL16.
Aoyama, T., Yamashita, S., Tomita, K.(2020) Nucleic Acids Res 48: 5157-5168
- PubMed: 32266935
- DOI: https://doi.org/10.1093/nar/gkaa227
- Primary Citation of Related Structures:
6M1U - PubMed Abstract:
The N6-methyladenosine modification at position 43 (m6A43) of U6 snRNA is catalyzed by METTL16, and is important for the 5'-splice site recognition by U6 snRNA during pre-mRNA splicing. Human METTL16 consists of the N-terminal methyltransferase domain (MTD) and the C-terminal vertebrate conserved region (VCR). While the MTD has an intrinsic property to recognize a specific sequence in the distinct structural context of RNA, the VCR functions have remained uncharacterized. Here, we present structural and functional analyses of the human METTL16 VCR. The VCR increases the affinity of METTL16 toward U6 snRNA, and the conserved basic region in VCR is important for the METTL16-U6 snRNA interaction. The VCR structure is topologically homologous to the C-terminal RNA binding domain, KA1, in U6 snRNA-specific terminal uridylyl transferase 1 (TUT1). A chimera of the N-terminal MTD of METTL16 and the C-terminal KA1 of TUT1 methylated U6 snRNA more efficiently than the MTD, indicating the functional conservation of the VCR and KA1 for U6 snRNA biogenesis. The VCR interacts with the internal stem-loop (ISL) within U6 snRNA, and this interaction would induce the conformational rearrangement of the A43-containing region of U6 snRNA, thereby modifying the RNA structure to become suitable for productive catalysis by the MTD. Therefore, the MTD and VCR in METTL16 cooperatively facilitate the m6A43 U6 snRNA modification.
Organizational Affiliation:
Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Kashiwa, Chiba 277-8562, Japan.