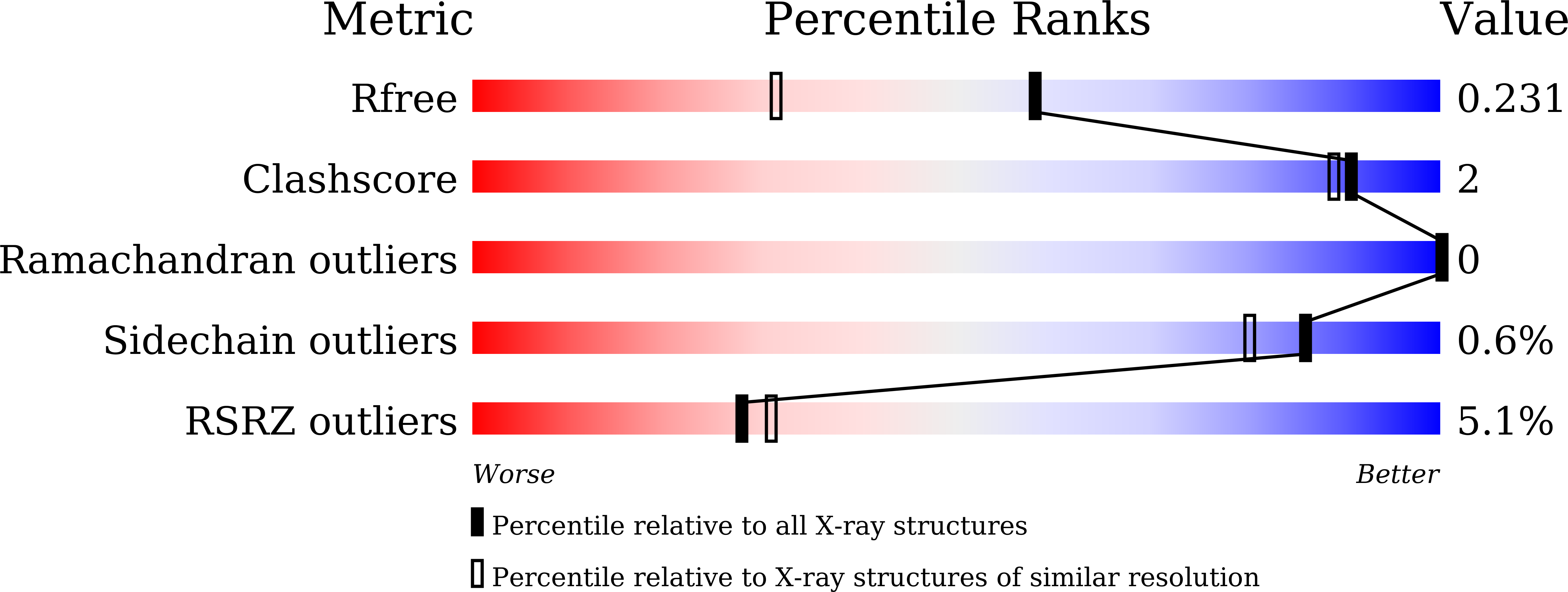
Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
Hu, Y., Shi, H., Zhou, M., Ren, Q., Zhu, W., Zhang, W., Zhang, Z., Zhou, C., Liu, Y., Ding, X., Shen, H.C., Yan, S.F., Dey, F., Wu, W., Zhai, G., Zhou, Z., Xu, Z., Ji, Y., Lv, H., Jiang, T., Wang, W., Xu, Y., Vercruysse, M., Yao, X., Mao, Y., Yu, X., Bradley, K., Tan, X.(2020) J Med Chem 63: 9623-9649
- PubMed: 32787097
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00768
- Primary Citation of Related Structures:
6M1J, 6M1S - PubMed Abstract:
The rise of multidrug resistant (MDR) Gram-negative (GN) pathogens and the decline of available antibiotics that can effectively treat these severe infections are a major threat to modern medicine. Developing novel antibiotics against MDR GN pathogens is particularly difficult as compounds have to permeate the GN double membrane, which has very different physicochemical properties, and have to circumvent a plethora of resistance mechanisms such as multiple efflux pumps and target modifications. The bacterial type II topoisomerases DNA gyrase (GyrA 2 B 2 ) and Topoisomerase IV (ParC 2 E 2 ) are highly conserved targets across all bacterial species and validated in the clinic by the fluoroquinolones. Dual inhibitors targeting the ATPase domains (GyrB/ParE) of type II topoisomerases can overcome target-based fluoroquinolone resistance. However, few ATPase inhibitors are active against GN pathogens. In this study, we demonstrated a successful strategy to convert a 2-carboxamide substituted azaindole chemical scaffold with only Gram-positive (GP) activity into a novel series with also potent activity against a range of MDR GN pathogens. By systematically fine-tuning the many physicochemical properties, we identified lead compounds such as 17r with a balanced profile showing potent GN activity, high aqueous solubility, and desirable PK features. Moreover, we showed the bactericidal efficacy of 17r using a neutropenic mouse thigh infection model.
Organizational Affiliation:
WuXi AppTec (Wuhan) Co., Ltd., No. 666 Gaoxin Road, Wuhan East Lake High-tech Development Zone, Hubei 430075, China.