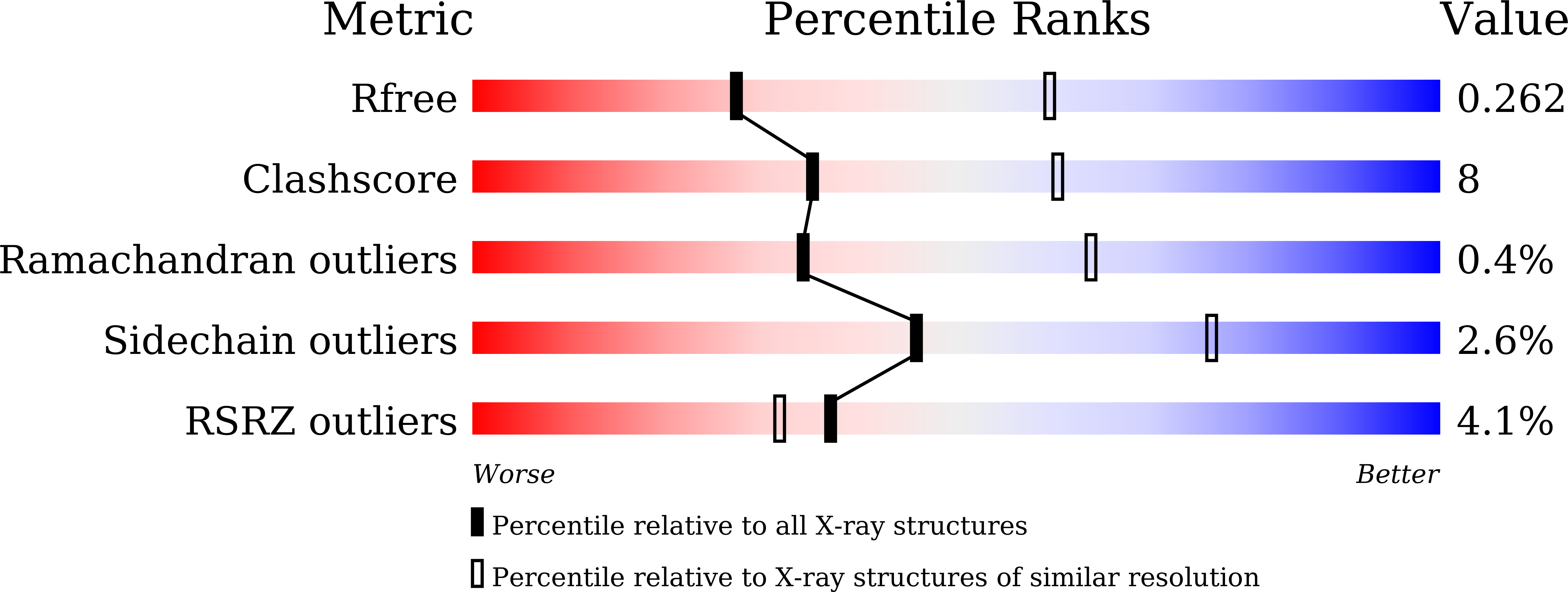
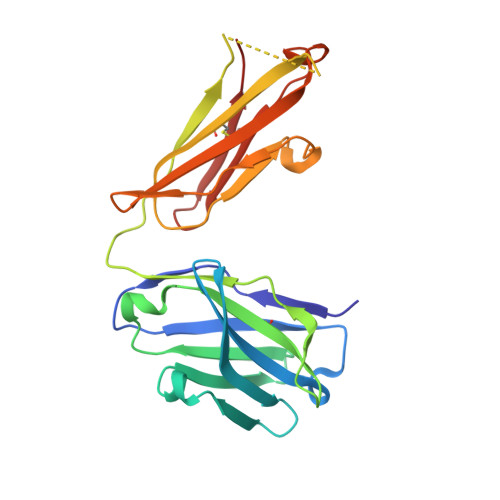
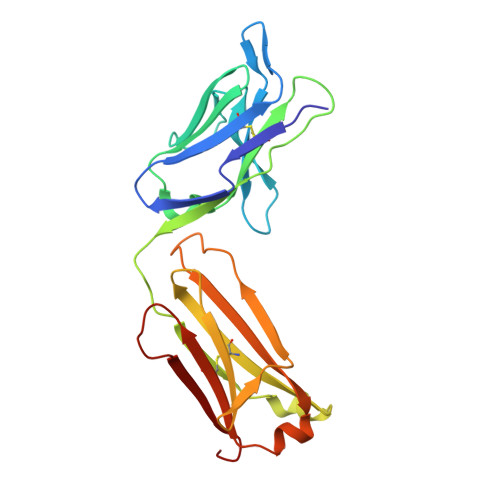
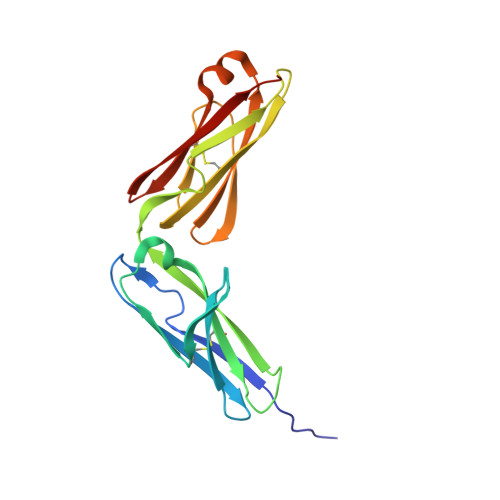
Structure basis for AA98 inhibition on the activation of endothelial cells mediated by CD146.
Chen, X., Yan, H., Liu, D., Xu, Q., Duan, H., Feng, J., Yan, X., Xie, C.(2021) iScience 24: 102417-102417
- PubMed: 33997697
- DOI: https://doi.org/10.1016/j.isci.2021.102417
- Primary Citation of Related Structures:
6LYN - PubMed Abstract:
CD146 is an adhesion molecule that plays important roles in angiogenesis, cancer metastasis, and immune response. It exists as a monomer or dimer on the cell surface. AA98 is a monoclonal antibody that binds to CD146, which abrogates the activation of CD146-mediated signaling pathways and shows inhibitory effects on tumor growth. However, how AA98 inhibits the function of CD146 remains unclear. Here, we describe a crystal structure of the CD146/AA98 Fab complex at a resolution of 2.8 Å. Monomeric CD146 is stabilized by AA98 Fab binding to the junction region of CD146 domains 4 and 5. A higher-affinity AA98 variant (here named HA98) was thus rationally designed. Better binding to CD146 and prominent inhibition on cell migration were achieved with HA98. Further experiments on xenografted melanoma in mice with HA98 revealed superior inhibitory effects on tumor growth to those of AA98, which suggested future applications of this antibody in cancer therapy.
Organizational Affiliation:
Key Laboratory of Protein and Peptide Pharmaceuticals, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.