Macrocyclic Peptide-Mediated Blockade of the CD47-SIRP alpha Interaction as a Potential Cancer Immunotherapy.
Hazama, D., Yin, Y., Murata, Y., Matsuda, M., Okamoto, T., Tanaka, D., Terasaka, N., Zhao, J., Sakamoto, M., Kakuchi, Y., Saito, Y., Kotani, T., Nishimura, Y., Nakagawa, A., Suga, H., Matozaki, T.(2020) Cell Chem Biol 27: 1181-1191.e7
- PubMed: 32640189
- DOI: https://doi.org/10.1016/j.chembiol.2020.06.008
- Primary Citation of Related Structures:
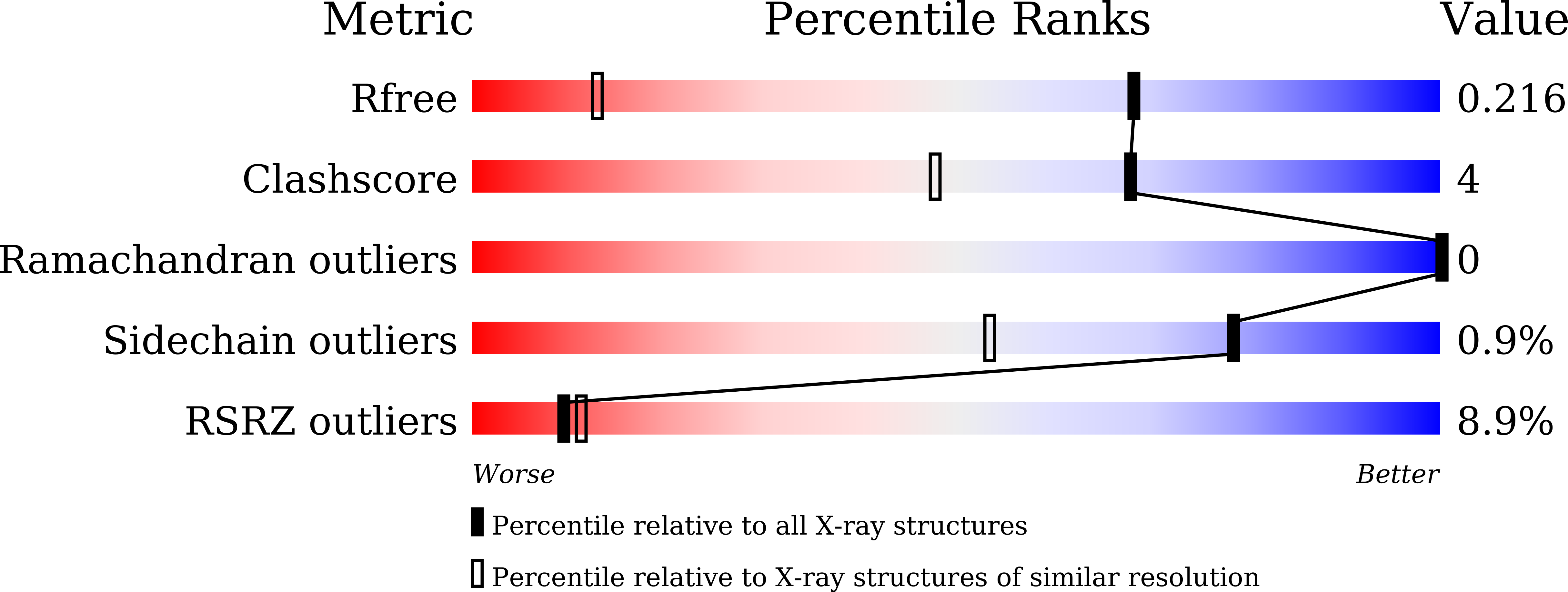
6LYC - PubMed Abstract:
Medium-sized macrocyclic peptides are an alternative to small compounds and large biomolecules as a class of pharmaceutics. The CD47-SIRPα signaling axis functions as an innate immune checkpoint that inhibits phagocytosis in phagocytes and has been implicated as a promising target for cancer immunotherapy. The potential of macrocyclic peptides that target this signaling axis as immunotherapeutic agents has remained unknown, however. Here we have developed a macrocyclic peptide consisting of 15 amino acids that binds to the ectodomain of mouse SIRPα and efficiently blocks its interaction with CD47 in an allosteric manner. The peptide markedly promoted the phagocytosis of antibody-opsonized tumor cells by macrophages in vitro as well as enhanced the inhibitory effect of anti-CD20 or anti-gp75 antibodies on tumor formation or metastasis in vivo. Our results suggest that allosteric inhibition of the CD47-SIRPα interaction by macrocyclic peptides is a potential approach to cancer immunotherapy.
Organizational Affiliation:
Division of Molecular and Cellular Signaling, Department of Biochemistry and Molecular Biology, Kobe University Graduate School of Medicine, Kobe 650-0017, Japan; Division of Respiratory Medicine, Department of Internal Medicine, Kobe University Graduate School of Medicine, Kobe 650-0017, Japan.