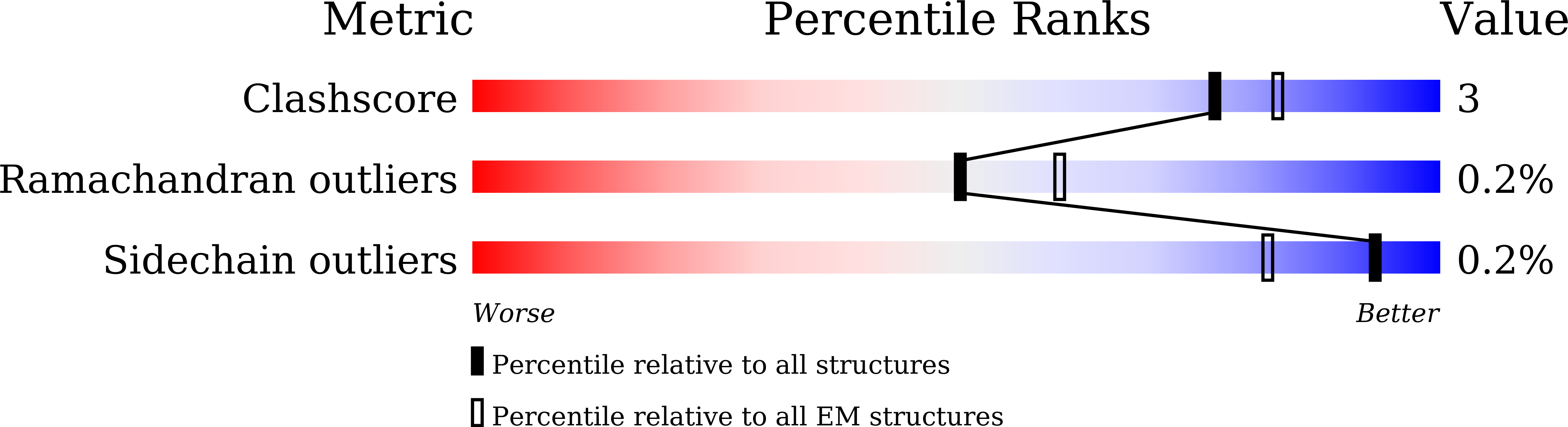
ClpL is a functionally active tetradecameric AAA+ chaperone, distinct from hexameric/dodecameric ones.
Kim, G., Lee, S.G., Han, S., Jung, J., Jeong, H.S., Hyun, J.K., Rhee, D.K., Kim, H.M., Lee, S.(2020) FASEB J 34: 14353-14370
- PubMed: 32910525
- DOI: https://doi.org/10.1096/fj.202000843R
- Primary Citation of Related Structures:
6LSY, 6LT4 - PubMed Abstract:
AAA+ (ATPases associated with diverse cellular activities) chaperones are involved in a plethora of cellular activities to ensure protein homeostasis. The function of AAA+ chaperones is mostly modulated by their hexameric/dodecameric quaternary structures. Here we report the structural and biochemical characterizations of a tetradecameric AAA+ chaperone, ClpL from Streptococcus pneumoniae. ClpL exists as a tetradecamer in solution in the presence of ATP. The cryo-EM structure of ClpL at 4.5 Å resolution reveals a striking tetradecameric arrangement. Solution structures of ClpL derived from small-angle X-ray scattering data suggest that the tetradecameric ClpL could assume a spiral conformation found in active hexameric/dodecameric AAA+ chaperone structures. Vertical positioning of the middle domain accounts for the head-to-head arrangement of two heptameric rings. Biochemical activity assays with site-directed mutagenesis confirmed the critical roles of residues both in the integrity of the tetradecameric arrangement and activities of ClpL. Non-conserved Q321 and R670 are crucial in the heptameric ring assembly of ClpL. These results establish that ClpL is a functionally active tetradecamer, clearly distinct from hexameric/dodecameric AAA+ chaperones.
Organizational Affiliation:
Department of Biological Sciences, Sungkyunkwan University, Suwon, Korea.