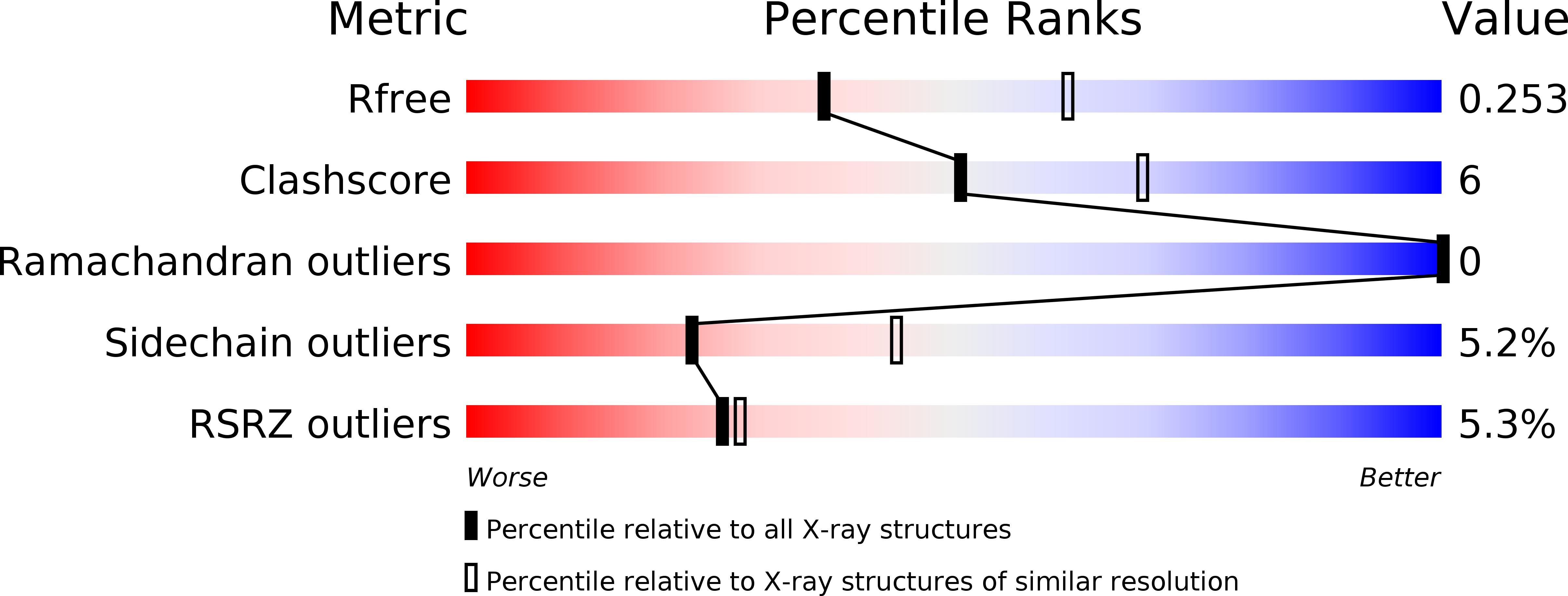
Structural insights into telomere protection and homeostasis regulation by yeast CST complex.
Ge, Y., Wu, Z., Chen, H., Zhong, Q., Shi, S., Li, G., Wu, J., Lei, M.(2020) Nat Struct Mol Biol 27: 752-762
- PubMed: 32661422
- DOI: https://doi.org/10.1038/s41594-020-0459-8
- Primary Citation of Related Structures:
6LBR, 6LBS, 6LBT, 6LBU - PubMed Abstract:
Budding yeast Cdc13-Stn1-Ten1 (CST) complex plays an essential role in telomere protection and maintenance. Despite extensive studies, only structural information of individual domains of CST is available; the architecture of CST still remains unclear. Here, we report crystal structures of Kluyveromyces lactis Cdc13-telomeric-DNA, Cdc13-Stn1 and Stn1-Ten1 complexes and propose an integrated model depicting how CST assembles and plays its roles at telomeres. Surprisingly, two oligonucleotide/oligosaccharide-binding (OB) folds of Cdc13 (OB2 and OB4), previously believed to mediate Cdc13 homodimerization, actually form a stable intramolecular interaction. This OB2-OB4 module of Cdc13 is required for the Cdc13-Stn1 interaction that assembles CST into an architecture with a central ring-like core and multiple peripheral modules in a 2:2:2 stoichiometry. Functional analyses indicate that this unique CST architecture is essential for both telomere capping and homeostasis regulation. Overall, our results provide fundamentally valuable structural information regarding the CST complex and its roles in telomere biology.
Organizational Affiliation:
State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China.