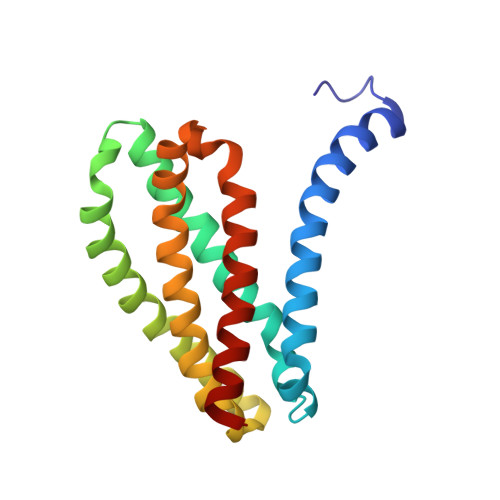
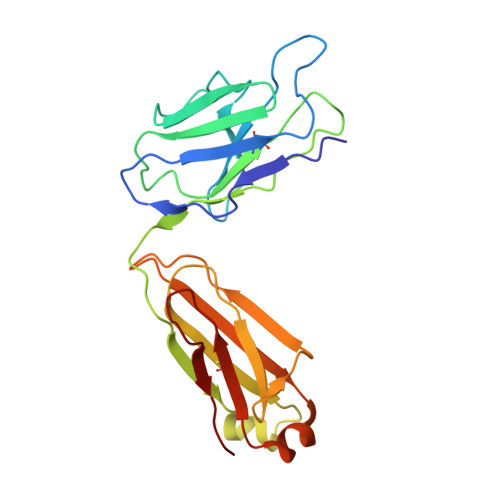
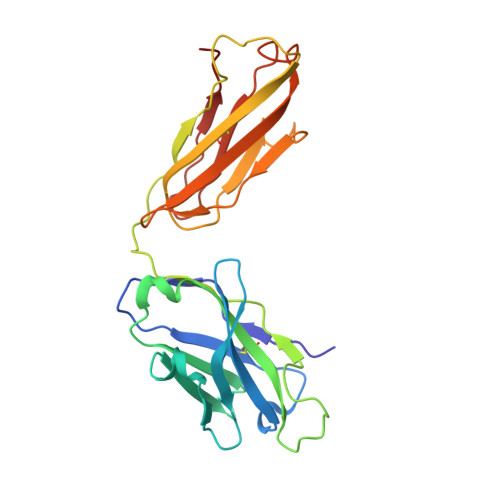
The structure of MgtE in the absence of magnesium provides new insights into channel gating.
Jin, F., Sun, M., Fujii, T., Yamada, Y., Wang, J., Maturana, A.D., Wada, M., Su, S., Ma, J., Takeda, H., Kusakizako, T., Tomita, A., Nakada-Nakura, Y., Liu, K., Uemura, T., Nomura, Y., Nomura, N., Ito, K., Nureki, O., Namba, K., Iwata, S., Yu, Y., Hattori, M.(2021) PLoS Biol 19: e3001231-e3001231
- PubMed: 33905418
- DOI: https://doi.org/10.1371/journal.pbio.3001231
- Primary Citation of Related Structures:
6LBH - PubMed Abstract:
MgtE is a Mg2+ channel conserved in organisms ranging from prokaryotes to eukaryotes, including humans, and plays an important role in Mg2+ homeostasis. The previously determined MgtE structures in the Mg2+-bound, closed-state, and structure-based functional analyses of MgtE revealed that the binding of Mg2+ ions to the MgtE cytoplasmic domain induces channel inactivation to maintain Mg2+ homeostasis. There are no structures of the transmembrane (TM) domain for MgtE in Mg2+-free conditions, and the pore-opening mechanism has thus remained unclear. Here, we determined the cryo-electron microscopy (cryo-EM) structure of the MgtE-Fab complex in the absence of Mg2+ ions. The Mg2+-free MgtE TM domain structure and its comparison with the Mg2+-bound, closed-state structure, together with functional analyses, showed the Mg2+-dependent pore opening of MgtE on the cytoplasmic side and revealed the kink motions of the TM2 and TM5 helices at the glycine residues, which are important for channel activity. Overall, our work provides structure-based mechanistic insights into the channel gating of MgtE.
Organizational Affiliation:
State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Shanghai Key Laboratory of Bioactive Small Molecules, Department of Physiology and Biophysics, School of Life Sciences, Fudan University, Shanghai, China.