Near-atomic resolution structures of interdigitated nucleosome fibres.
Adhireksan, Z., Sharma, D., Lee, P.L., Davey, C.A.(2020) Nat Commun 11: 4747-4747
- PubMed: 32958761
- DOI: https://doi.org/10.1038/s41467-020-18533-2
- Primary Citation of Related Structures:
6LA8, 6LA9, 6M3V, 6M44 - PubMed Abstract:
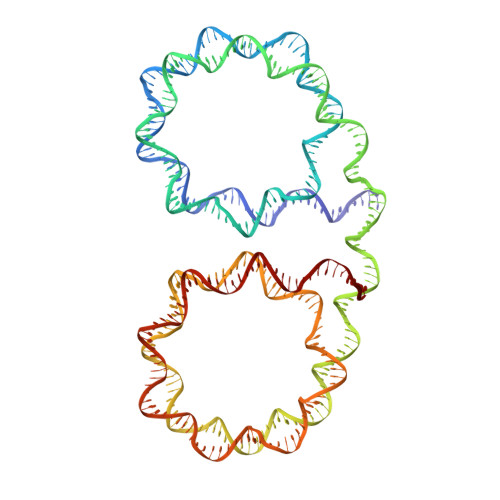
Chromosome structure at the multi-nucleosomal level has remained ambiguous in spite of its central role in epigenetic regulation and genome dynamics. Recent investigations of chromatin architecture portray diverse modes of interaction within and between nucleosome chains, but how this is realized at the atomic level is unclear. Here we present near-atomic resolution crystal structures of nucleosome fibres that assemble from cohesive-ended dinucleosomes with and without linker histone. As opposed to adopting folded helical '30 nm' structures, the fibres instead assume open zigzag conformations that are interdigitated with one another. Zigzag conformations obviate extreme bending of the linker DNA, while linker DNA size (nucleosome repeat length) dictates fibre configuration and thus fibre-fibre packing, which is supported by variable linker histone binding. This suggests that nucleosome chains have a predisposition to interdigitate with specific characteristics under condensing conditions, which rationalizes observations of local chromosome architecture and the general heterogeneity of chromatin structure.
Organizational Affiliation:
School of Biological Sciences, Nanyang Technological University, 60 Nanyang Drive, Singapore, 637551, Singapore.