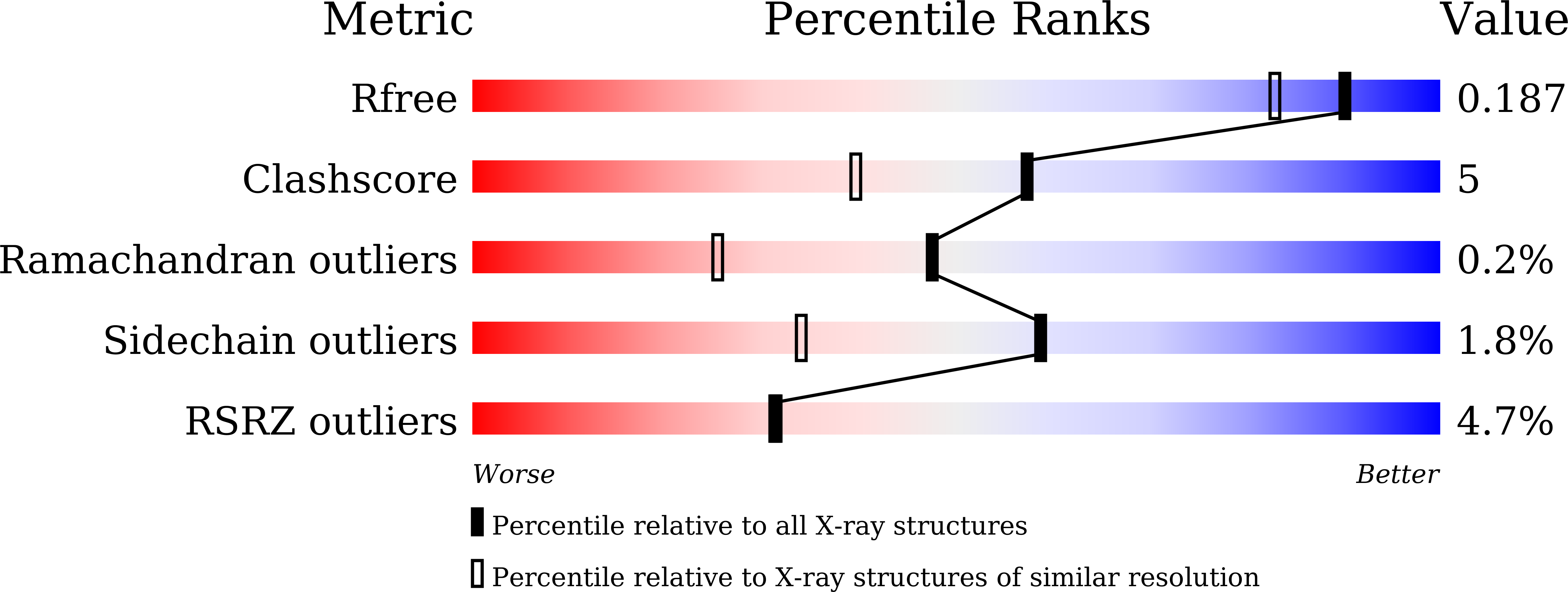
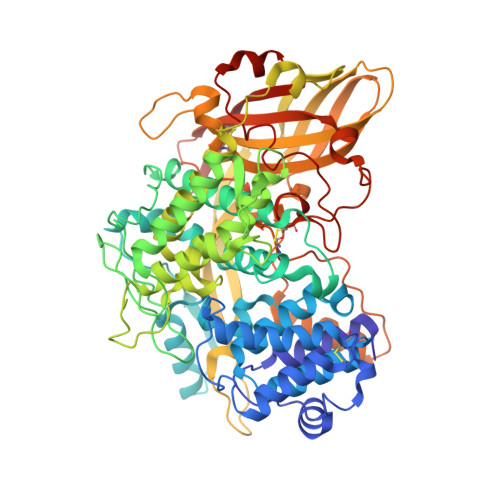
The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase.
Masuda, T., Baba, S., Matsuo, K., Ito, S., Mikami, B.(2020) Arch Biochem Biophys 688: 108370-108370
- PubMed: 32380017
- DOI: https://doi.org/10.1016/j.abb.2020.108370
- Primary Citation of Related Structures:
6L8S - PubMed Abstract:
Hemocyanin (Hc) and phenoloxidase (PO) are members of the type 3 copper protein family. Although arthropod Hc and PO exhibit similar three-dimensional structures of the copper-containing active site, Hc functions as an oxygen transport protein, showing minimal or no phenoloxidase activity. Here, we present the crystal structure of the oxy form of Hc from Panulirus japonicus (PjHc) at 1.58 Å resolution. The structure of the di-copper active site of PjHc was found to be almost identical to that of PO. Although conserved amino acids and the water molecule crucial for the enzymatic activity were observed in PjHc at almost the same positions as those in PO, PjHc showed no enzymatic activity under our experimental conditions. One striking difference between PjHc and arthropod PO was the presence of a "blocker residue" near the binuclear copper site of PjHc. This blocker residue comprised a phenylalanine residue tightly stacked with an imidazole ring of a CuA coordinated histidine and hindered substrates from accessing the active site. Our results suggest that the blocker residue is also a determining factor of the catalytic activity of type 3 copper proteins.
Organizational Affiliation:
Laboratory of Marine Biology, Division of Applied Biological Science, Faculty of Agriculture, Setsunan University, 45-1 Nagaotoge-cho, Hirakata, Osaka, 573-0101, Japan. Electronic address: taro.masuda@setsunan.ac.jp.