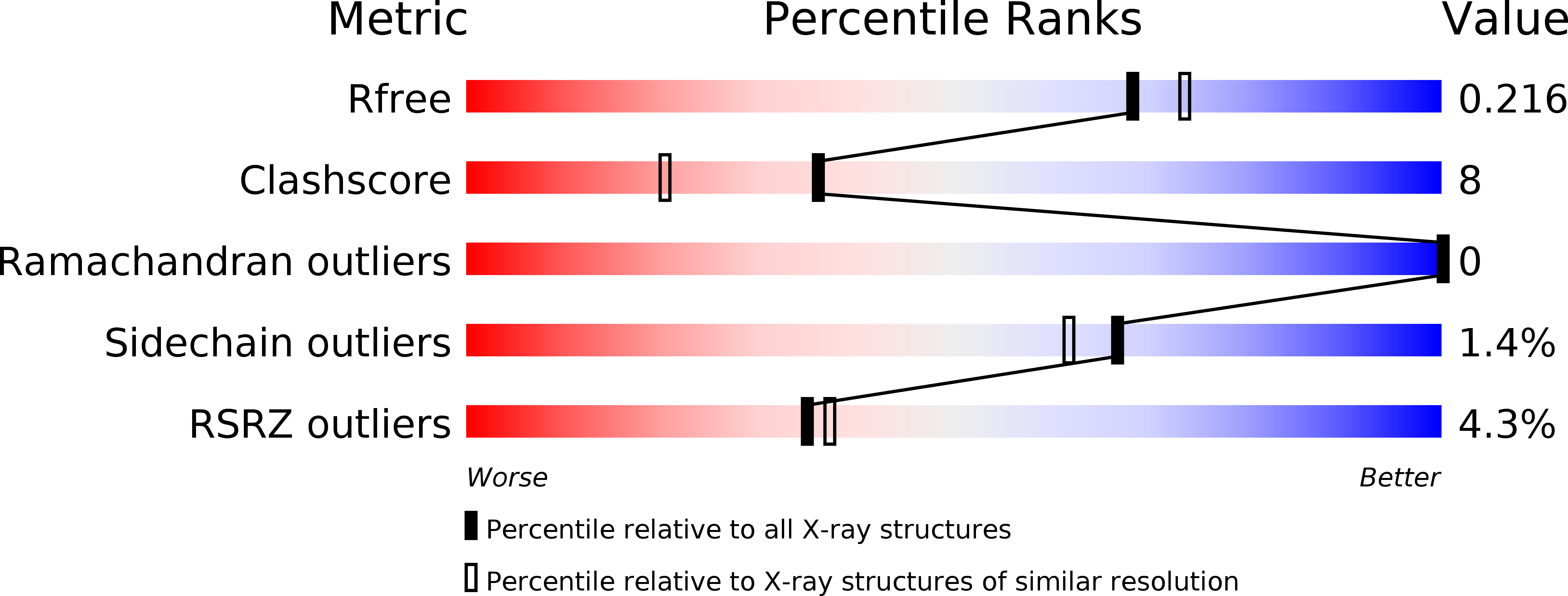
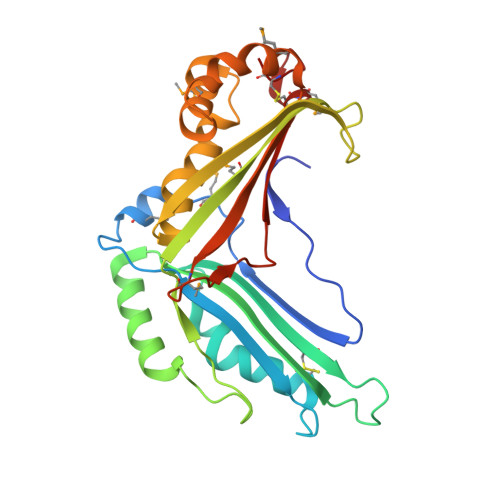
Crystal structure of a hypothetical T2SS effector Lpg0189 from Legionella pneumophila reveals a novel protein fold.
Chen, X., Liu, S., Jiang, S., Zhang, X., Zhang, N., Ma, J., Ge, H.(2020) Biochem Biophys Res Commun 521: 799-805
- PubMed: 31706575
- DOI: https://doi.org/10.1016/j.bbrc.2019.10.195
- Primary Citation of Related Structures:
6L6G, 6L6H - PubMed Abstract:
Lpg0189 is a type II secretion system-dependent extracellular protein with unknown function from Legionella pneumophila. Herein, we determined the crystal structure of Lpg0189 at 1.98 Å resolution by using single-wavelength anomalous diffraction (SAD). Lpg0189 folds into a novel chair-shaped architecture, with two sheets roughly perpendicular to each other. Bioinformatics analysis suggests Lpg0189 and its homologues are unique to Legionellales and evolved divergently. The interlinking structural and bioinformatics studies provide a better understanding of this hypothetical protein.
Organizational Affiliation:
Institute of Physical Science and Information Technology, Anhui University, Hefei, Anhui, 230601, China; School of Chemistry and Chemical Engineering, Anhui University, Hefei, Anhui, 230601, China.