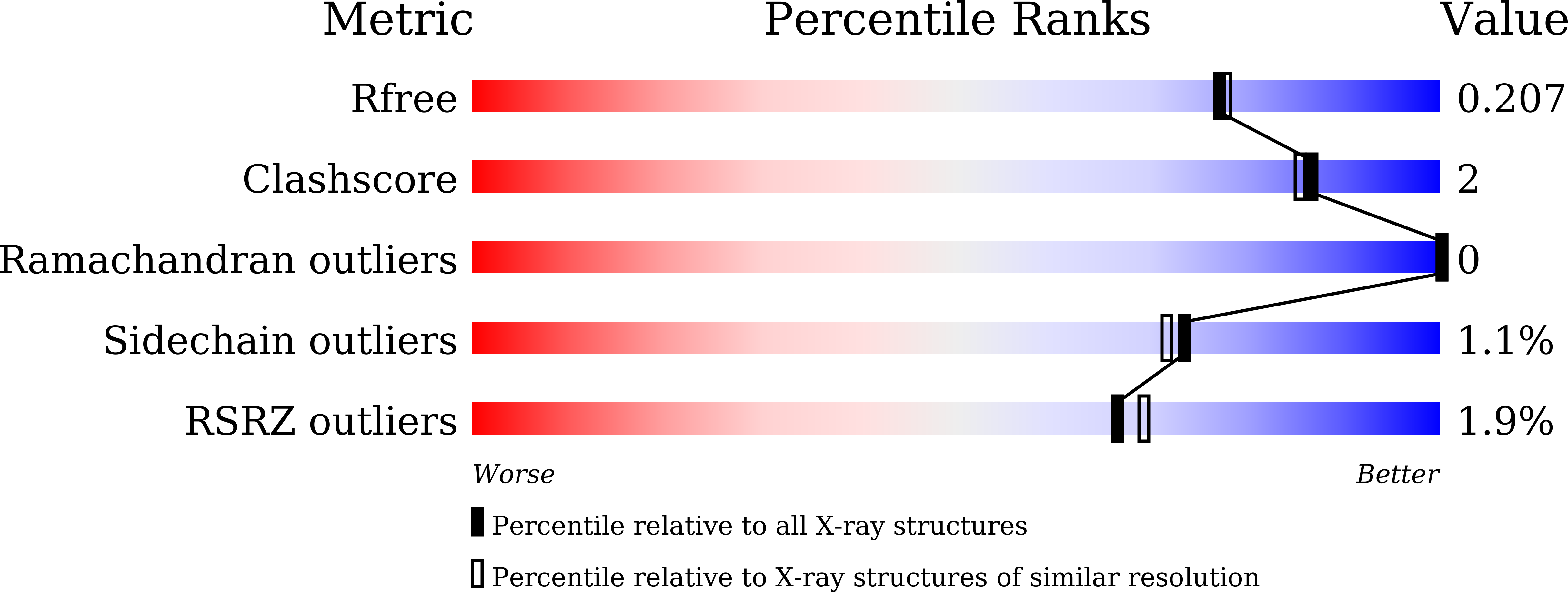
Structure of Human Phosphodiesterase 5A1 Complexed with Avanafil Reveals Molecular Basis of Isoform Selectivity and Guidelines for Targeting alpha-Helix Backbone Oxygen by Halogen Bonding.
Hsieh, C.M., Chen, C.Y., Chern, J.W., Chan, N.L.(2020) J Med Chem 63: 8485-8494
- PubMed: 32663396
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00853
- Primary Citation of Related Structures:
6L6E - PubMed Abstract:
Phosphodiesterase 5A1 (PDE5) is a key target for treating cardiovascular diseases and erectile dysfunction. Here, we report the crystal structure of PDE5 complexed with the sole second generation drug avanafil. Analysis of protein-drug interactions revealed the structural basis of avanafil's superior isoform selectivity. Moreover, a halogen bonding was observed between avanafil and a backbone carbonyl oxygen of an adjacent α-helix, whose contribution to inhibitory potency illustrates the feasibility of exploiting α-helix backbone in structure-based drug design.
Organizational Affiliation:
Institute of Biochemistry and Molecular Biology, College of Medicine, National Taiwan University, Taipei 100, Taiwan.