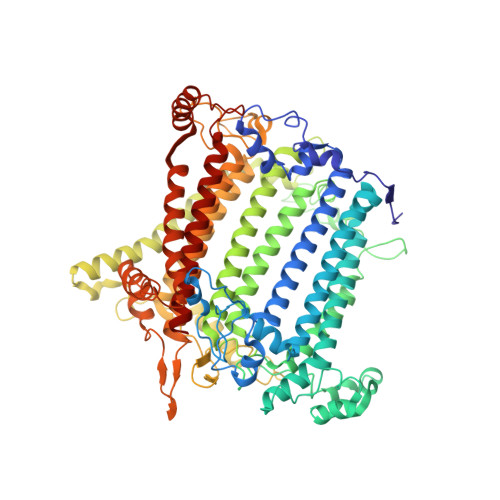
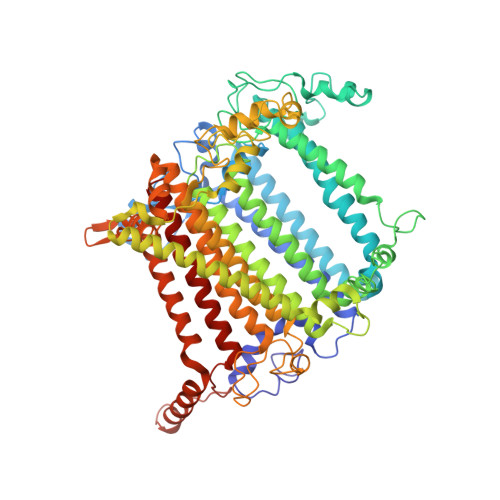
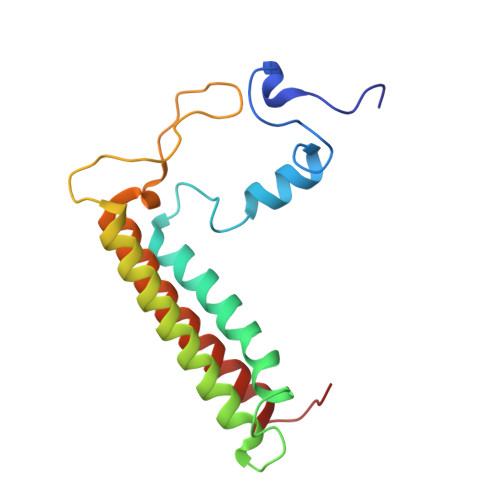
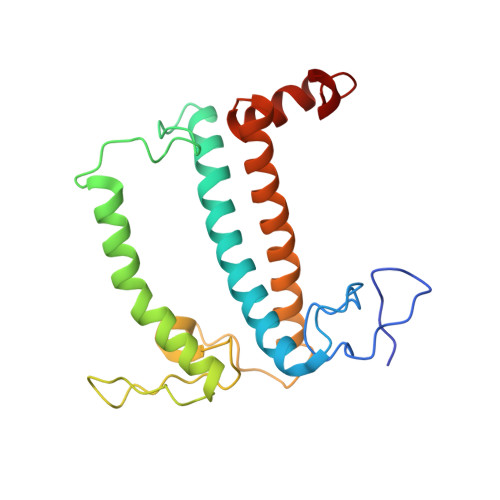
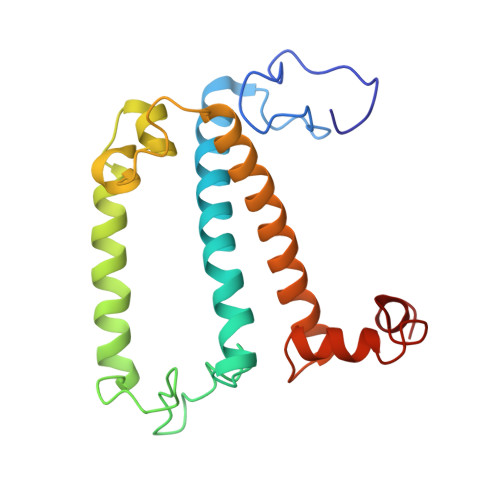
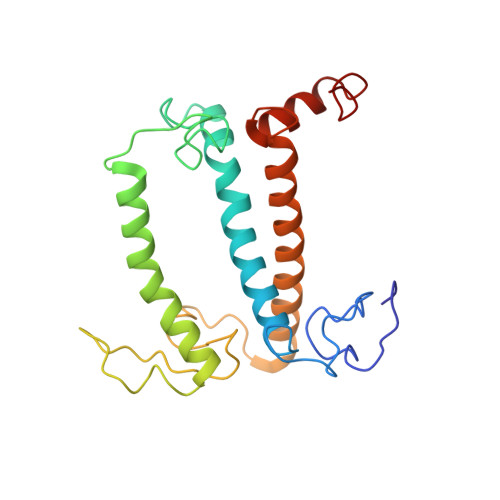
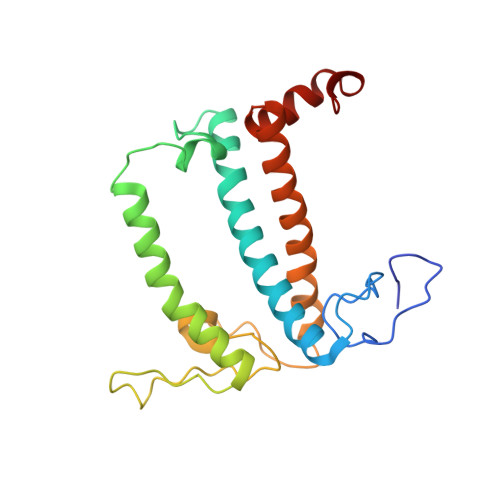
Antenna arrangement and energy-transfer pathways of PSI-LHCI from the moss Physcomitrella patens.
Yan, Q., Zhao, L., Wang, W., Pi, X., Han, G., Wang, J., Cheng, L., He, Y.K., Kuang, T., Qin, X., Sui, S.F., Shen, J.R.(2021) Cell Discov 7: 10-10
- PubMed: 33589616
- DOI: https://doi.org/10.1038/s41421-021-00242-9
- Primary Citation of Related Structures:
6L35 - PubMed Abstract:
Plants harvest light energy utilized for photosynthesis by light-harvesting complex I and II (LHCI and LHCII) surrounding photosystem I and II (PSI and PSII), respectively. During the evolution of green plants, moss is at an evolutionarily intermediate position from aquatic photosynthetic organisms to land plants, being the first photosynthetic organisms that landed. Here, we report the structure of the PSI-LHCI supercomplex from the moss Physcomitrella patens (Pp) at 3.23 Å resolution solved by cryo-electron microscopy. Our structure revealed that four Lhca subunits are associated with the PSI core in an order of Lhca1-Lhca5-Lhca2-Lhca3. This number is much decreased from 8 to 10, the number of subunits in most green algal PSI-LHCI, but the same as those of land plants. Although Pp PSI-LHCI has a similar structure as PSI-LHCI of land plants, it has Lhca5, instead of Lhca4, in the second position of Lhca, and several differences were found in the arrangement of chlorophylls among green algal, moss, and land plant PSI-LHCI. One chlorophyll, PsaF-Chl 305, which is found in the moss PSI-LHCI, is located at the gap region between the two middle Lhca subunits and the PSI core, and therefore may make the excitation energy transfer from LHCI to the core more efficient than that of land plants. On the other hand, energy-transfer paths at the two side Lhca subunits are relatively conserved. These results provide a structural basis for unravelling the mechanisms of light-energy harvesting and transfer in the moss PSI-LHCI, as well as important clues on the changes of PSI-LHCI after landing.
Organizational Affiliation:
Photosynthesis Research Center, Key Laboratory of Photobiology, Institute of Botany, Chinese Academy of Sciences, Beijing, 100093, China.