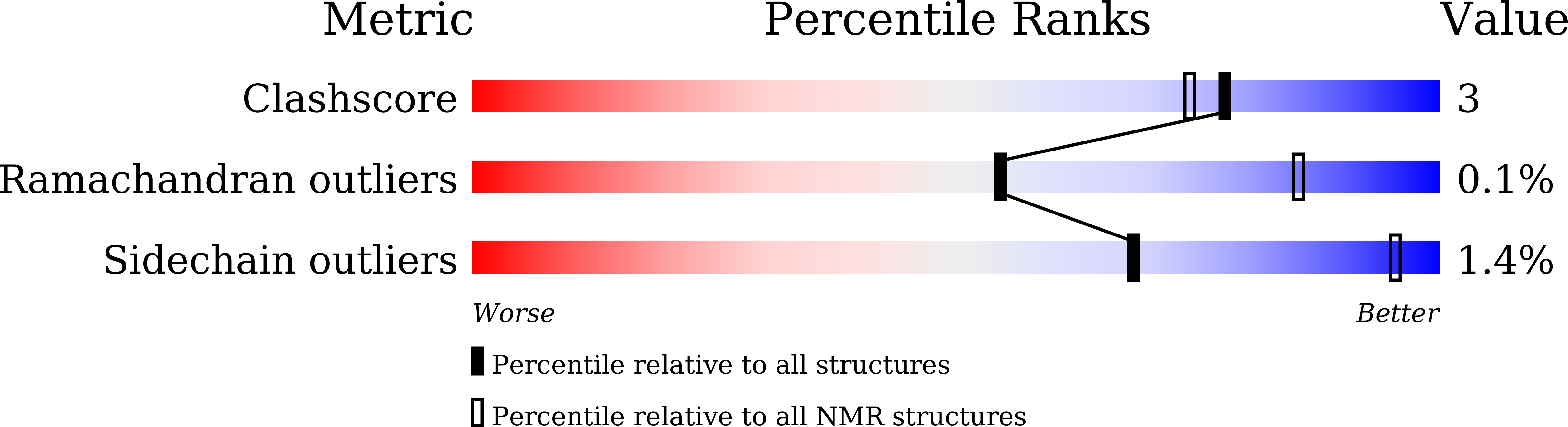Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Li, R.X., Zhang, N.N., Wu, B., OuYang, B., Shen, H.B.(2021) Comput Struct Biotechnol J 19: 2575-2587
- PubMed: 34025944
- DOI: https://doi.org/10.1016/j.csbj.2021.04.046
- Primary Citation of Related Structures:
6L0L - PubMed Abstract:
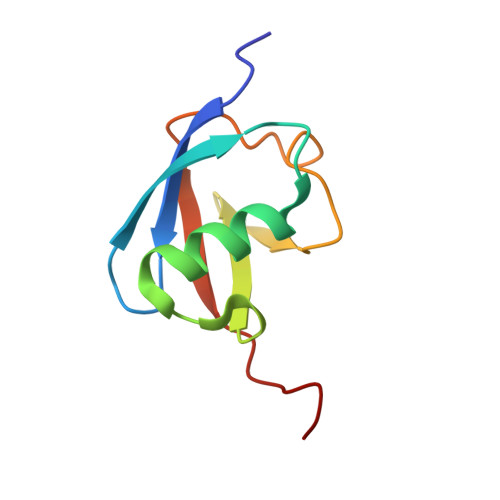
Protein design usually involves sequence search process and evaluation criteria. Commonly used methods primarily implement the Monte Carlo or simulated annealing algorithm with a single-energy function to obtain ideal solutions, which is often highly time-consuming and limited by the accuracy of the energy function. In this report, we introduce a multiobjective algorithm named Hydra for protein design, which employs two different energy functions to optimize solutions simultaneously and makes use of the latent quantitative relationship between different amino acid types to facilitate the search process. The framework uses two kinds of prior information to transform the original disordered discrete sequence space into a relatively ordered space, and decoy sequences are searched in this ordered space through a multiobjective swarm intelligence algorithm. This algorithm features high accuracy and a high-speed search process. Our method was tested on 40 targets covering different fold classes, which were computationally verified to be well folded, and it experimentally solved the 1UBQ fold by NMR in excellent agreement with the native structure with a backbone RMSD deviation of 1.074 Å. The Hydra software package can be downloaded from: http://www.csbio.sjtu.edu.cn/bioinf/HYDRA/ for academic use.
Organizational Affiliation:
Institute of Image Processing and Pattern Recognition, Shanghai Jiao Tong University, and Key Laboratory of System Control and Information Processing, Ministry of Education of China, Shanghai 200240, China.