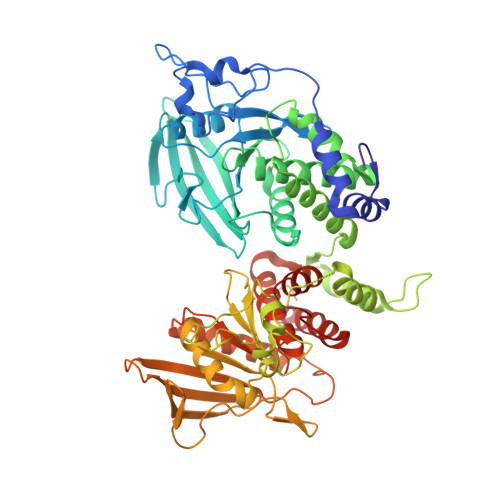
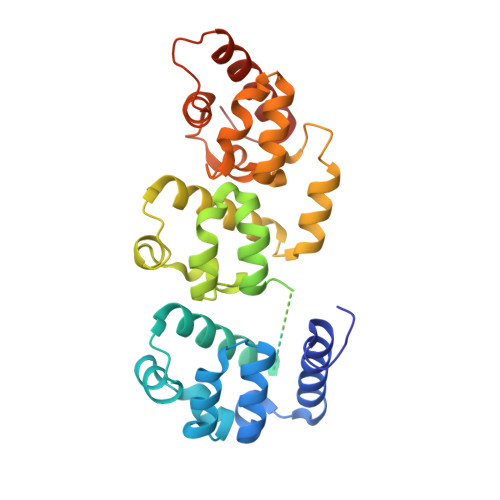
Structural basis of liprin-alpha-promoted LAR-RPTP clustering for modulation of phosphatase activity.
Xie, X., Luo, L., Liang, M., Zhang, W., Zhang, T., Yu, C., Wei, Z.(2020) Nat Commun 11: 169-169
- PubMed: 31924785
- DOI: https://doi.org/10.1038/s41467-019-13949-x
- Primary Citation of Related Structures:
6KR4 - PubMed Abstract:
Leukocyte common antigen-related receptor protein tyrosine phosphatases (LAR-RPTPs) are cell adhesion molecules involved in mediating neuronal development. The binding of LAR-RPTPs to extracellular ligands induces local clustering of LAR-RPTPs to regulate axon growth and synaptogenesis. LAR-RPTPs interact with synaptic liprin-α proteins via the two cytoplasmic phosphatase domains, D1 and D2. Here we solve the crystal structure of LAR_D1D2 in complex with the SAM repeats of liprin-α3, uncovering a conserved two-site binding mode. Cellular analysis shows that liprin-αs robustly promote clustering of LAR in cells by both the liprin-α/LAR interaction and the oligomerization of liprin-α. Structural analysis reveals a unique homophilic interaction of LAR via the catalytically active D1 domains. Disruption of the D1/D1 interaction diminishes the liprin-α-promoted LAR clustering and increases tyrosine dephosphorylation, demonstrating that the phosphatase activity of LAR is negatively regulated by forming clusters. Additionally, we find that the binding of LAR to liprin-α allosterically regulates the liprin-α/liprin-β interaction.
Organizational Affiliation:
Department of Biology, Southern University of Science and Technology, Shenzhen, Guangdong, 518055, China.