Substrates promiscuity of xyloglucanases and endoglucanases of glycoside hydrolase 12 family
Hong, Y., Tao, T., Pengjun, S., Jiaming, C., Xiaoyu, W., Chen, H., yingguo, B., Bin, Y.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| GH12 beta-1, 4-endoglucanase | 219 | Aspergillus fischeri | Mutation(s): 0 Gene Names: Nfeg12A EC: 3.2.1.4 | 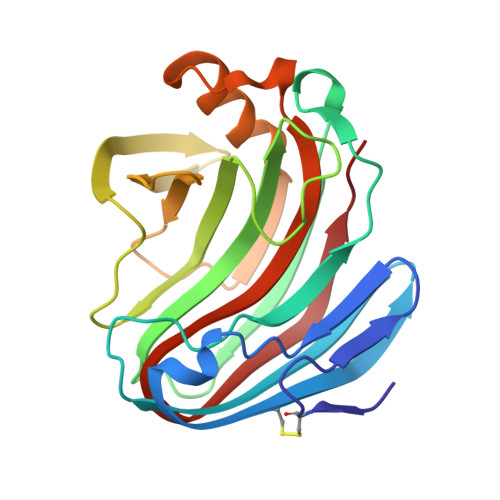 | |
UniProt | |||||
Find proteins for A0A1L6CE30 (Aspergillus fischeri) Explore A0A1L6CE30 Go to UniProtKB: A0A1L6CE30 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A1L6CE30 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 92.708 | α = 90 |
| b = 92.708 | β = 90 |
| c = 119.928 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Coot | model building |
| PHASER | phasing |