Chaperone-tip adhesin complex is vital for synergistic activation of CFA/I fimbriae biogenesis.
He, L.H., Wang, H., Liu, Y., Kang, M., Li, T., Li, C.C., Tong, A.P., Zhu, Y.B., Song, Y.J., Savarino, S.J., Prouty, M.G., Xia, D., Bao, R.(2020) PLoS Pathog 16: e1008848-e1008848
- PubMed: 33007034
- DOI: https://doi.org/10.1371/journal.ppat.1008848
- Primary Citation of Related Structures:
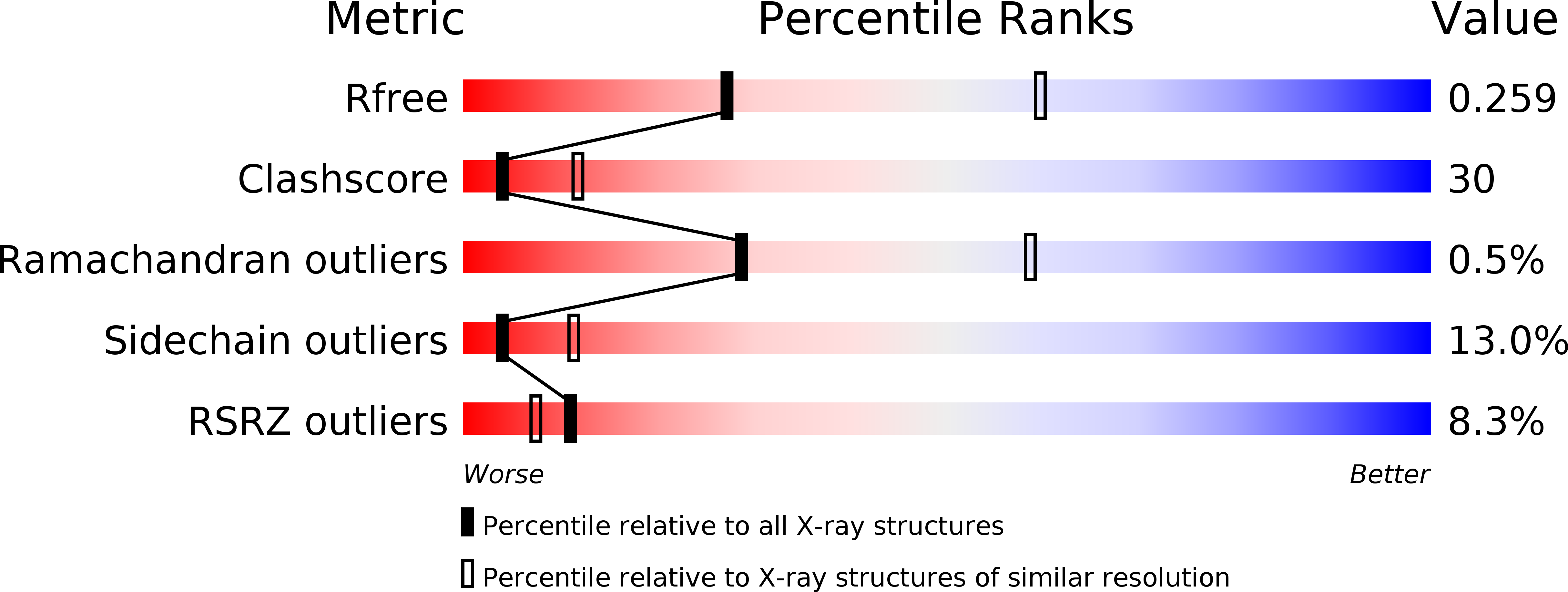
6K73 - PubMed Abstract:
Colonization factor CFA/I defines the major adhesive fimbriae of enterotoxigenic Escherichia coli and mediates bacterial attachment to host intestinal epithelial cells. The CFA/I fimbria consists of a tip-localized minor adhesive subunit, CfaE, and thousands of copies of the major subunit CfaB polymerized into an ordered helical rod. Biosynthesis of CFA/I fimbriae requires the assistance of the periplasmic chaperone CfaA and outer membrane usher CfaC. Although the CfaE subunit is proposed to initiate the assembly of CFA/I fimbriae, how it performs this function remains elusive. Here, we report the establishment of an in vitro assay for CFA/I fimbria assembly and show that stabilized CfaA-CfaB and CfaA-CfaE binary complexes together with CfaC are sufficient to drive fimbria formation. The presence of both CfaA-CfaE and CfaC accelerates fimbria formation, while the absence of either component leads to linearized CfaB polymers in vitro. We further report the crystal structure of the stabilized CfaA-CfaE complex, revealing features unique for biogenesis of Class 5 fimbriae.
Organizational Affiliation:
Center of Infectious Diseases, State Key Laboratory of Biotherapy, West China Hospital, Sichuan University, Chengdu, China.