Structural insights into the activation of USP46 by WDR48 and WDR20.
Zhu, H., Zhang, T., Wang, F., Yang, J., Ding, J.(2019) Cell Discov 5: 34-34
- PubMed: 31632687
- DOI: https://doi.org/10.1038/s41421-019-0102-1
- Primary Citation of Related Structures:
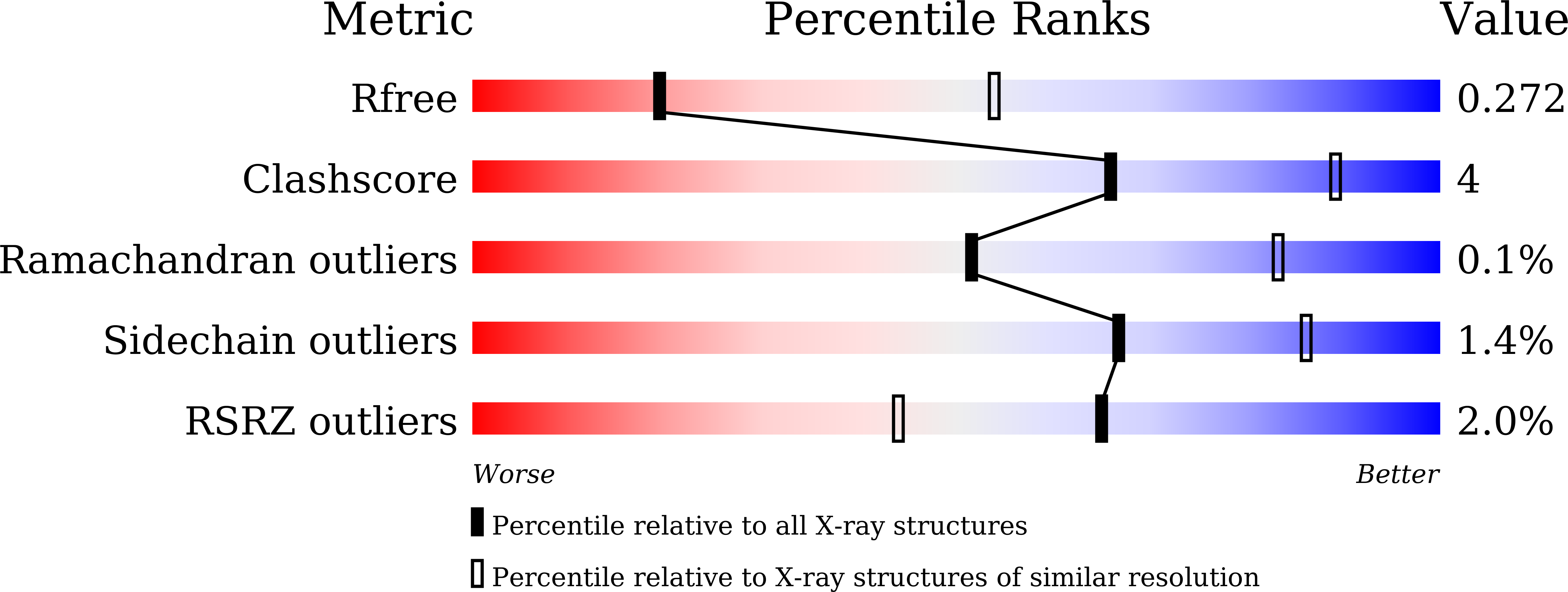
6JLQ
Organizational Affiliation:
State Key Laboratory of Molecular Biology, CAS Center for Excellence in Molecular Cell Science, Shanghai Institute of Biochemistry and Cell Biology, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yue-Yang Road, Shanghai, 200031 China.