Structure-Function Analyses of a Keratin Heterotypic Complex Identify Specific Keratin Regions Involved in Intermediate Filament Assembly.
Lee, C.H., Kim, M.S., Li, S., Leahy, D.J., Coulombe, P.A.(2020) Structure 28: 355-362.e4
- PubMed: 31995743
- DOI: https://doi.org/10.1016/j.str.2020.01.002
- Primary Citation of Related Structures:
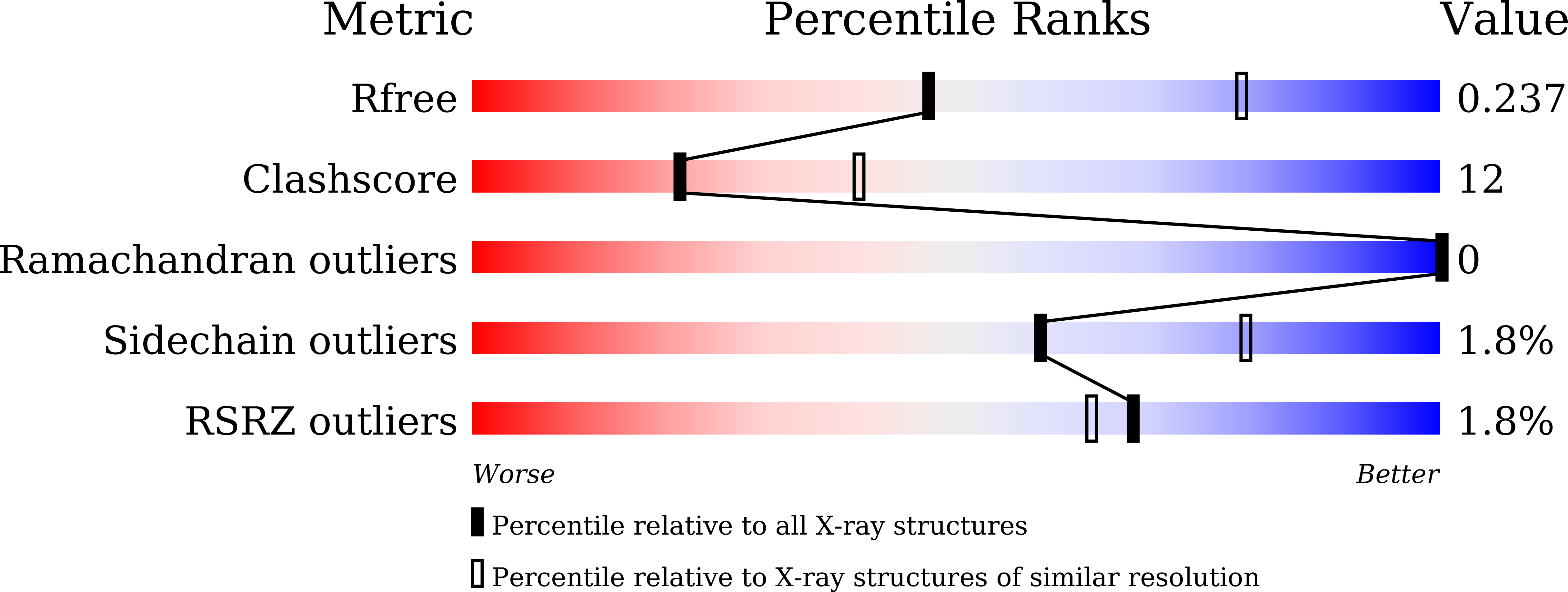
6JFV - PubMed Abstract:
Intermediate filaments (IFs) provide vital mechanical support in a broad array of cell types. Interference with this role causes cell fragility and accounts for a large number of human diseases. Gaining an understanding of the structure of IFs is paramount to understanding their function and designing therapeutic agents for relevant diseases. Here, we report the 2.6-Å resolution crystal structure of a complex of interacting 2B domains of keratin 5 (K5) and K14. K5 and K14 form a long-range, left-handed coiled coil, with participating α helices aligned in parallel and in register. Follow-up mutagenesis revealed that specific contacts between interacting 2B domains play a crucial role during 10-nm IF assembly, likely at the step of octamer-octamer association. The resulting structural model represents an atomic-resolution visualization of 2B-2B interactions important to filament assembly and provides insight into the defects introduced by mutations in IF genes associated with human skin diseases.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA.