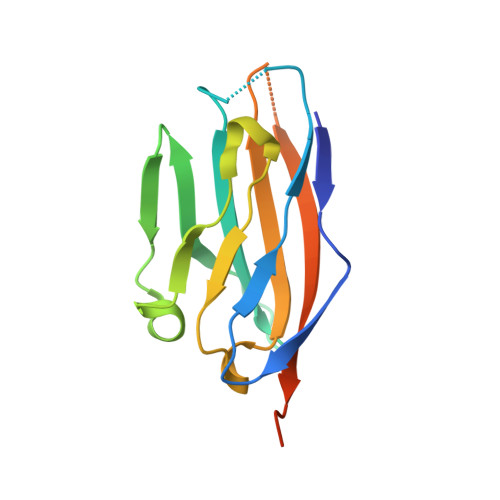
Structural and biochemical studies of the extracellular domain of Myelin protein zero-like protein 1
Yu, T., Liang, L., Zhao, X., Yin, Y.(2018) Biochem Biophys Res Commun 506: 883-890
- PubMed: 30392906
- DOI: https://doi.org/10.1016/j.bbrc.2018.10.161
- Primary Citation of Related Structures:
6IGO, 6IGT, 6IGW - PubMed Abstract:
Myelin protein zero-like protein 1 (MPZL1) is a member of the immunoglobulin superfamily, and is also a receptor of concanavalin A (ConA). MPZL1 is upregulated in hepatocellular carcinoma (HCC) and accelerates migration of HCC cells. However, function of MPZL1 as a receptor of ConA and its role in HCC development are largely unknown. To elucidate the functional basis, we have determined the crystal structure of the extracellular domain of MPZL1 at 2.7 Å resolution. Overall, it folds like a typical immunoglobulin variable-like domain that is much like MPZ. Unexpectedly, we found Asn50 is a unique glycosylation site and the glycosylation mediates its interaction with ConA. Furthermore, we also found that MPZL1 exists as a homodimer in the crystal, in which hydrogen bonds between Ser86 and Val145 play an important role. Our results demonstrate that glycosylation of Asn50 is essential for its function as a receptor of ConA. We propose that dimerization of MPZL1 participates in control of its signal transmission in cell adhesion.
Organizational Affiliation:
Peking-Tsinghua Center for Life Sciences, Peking University, Beijing, 100871, China.