Inhibition Mechanism of Urease by Au(III) Compounds Unveiled by X-ray Diffraction Analysis.
Mazzei, L., Wenzel, M.N., Cianci, M., Palombo, M., Casini, A., Ciurli, S.(2019) ACS Med Chem Lett 10: 564-570
- PubMed: 30996797
- DOI: https://doi.org/10.1021/acsmedchemlett.8b00585
- Primary Citation of Related Structures:
6I9Y - PubMed Abstract:
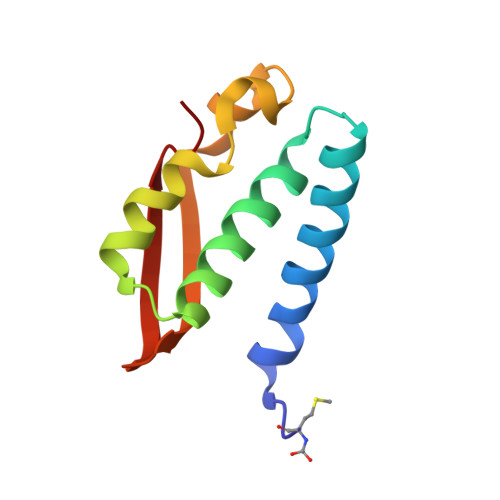
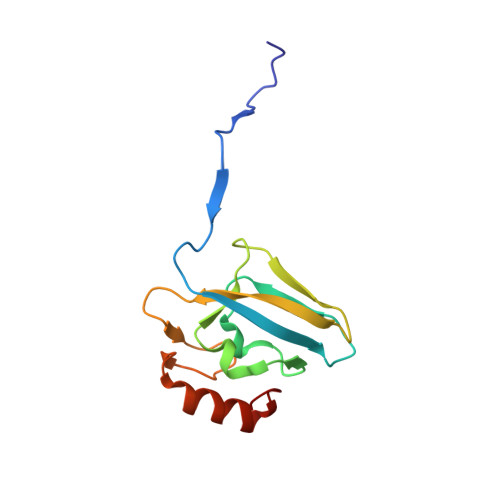
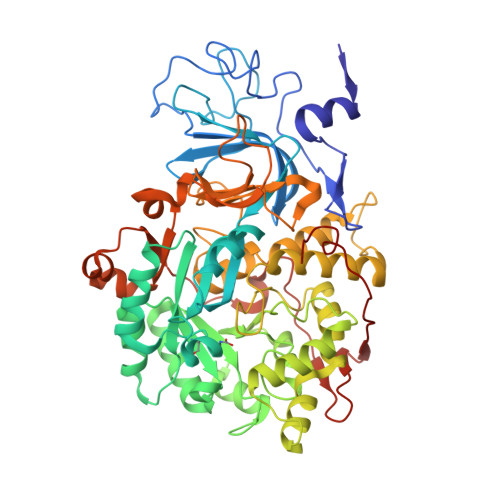
The nickel-dependent enzyme urease is a virulence factor for a large number of critical human pathogens, making this enzyme a potential target of therapeutics for the treatment of resistant bacterial infections. In the search for novel urease inhibitors, five selected coordination and organometallic Au(III) compounds containing N ∧ N or C ∧ N and C ∧ N ∧ N ligands were tested for their inhibitory effects against Canavalia ensiformis (jack bean) urease. The results showed potent inhibition effects with IC 50 values in the nanomolar range. The 2.14 Å resolution crystal structure of Sporosarcina pasteurii urease inhibited by the most effective Au(III) compound [Au(PbImMe)Cl 2 ]PF 6 (PbImMe = 1-methyl-2-(pyridin-2-yl)-benzimidazole) reveals the presence of two Au ions bound to the conserved triad αCys322/αHis323/αMet367. The binding of the Au ions to these residues blocks the movement of a flap, located at the edge of the active site channel and essential for enzyme catalysis, completely obliterating the catalytic activity of urease. Overall, the obtained results constitute the basis for the design of new gold complexes as selective urease inhibitors with future antibacterial applications.
Organizational Affiliation:
Laboratory of Bioinorganic Chemistry, Department of Pharmacy and Biotechnology, University of Bologna, Viale Giuseppe Fanin 40, I-40127 Bologna, Italy.