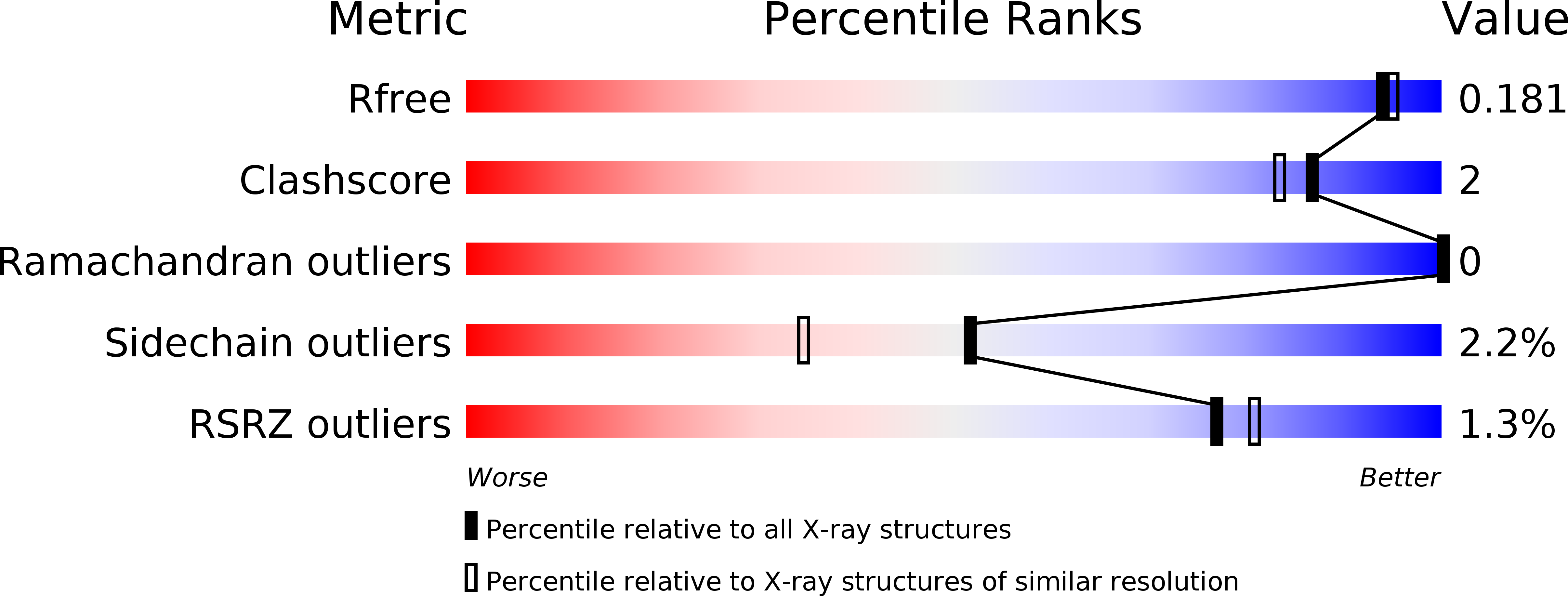
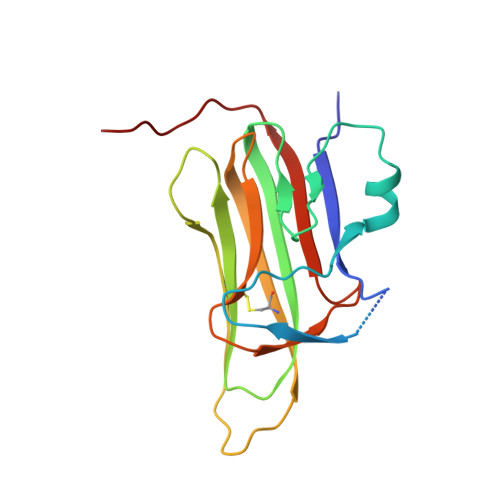
High-resolution crystal structure of arthropod Eiger TNF suggests a mode of receptor engagement and altered surface charge within endosomes.
Bertinelli, M., Paesen, G.C., Grimes, J.M., Renner, M.(2019) Commun Biol 2: 293-293
- PubMed: 31396573
- DOI: https://doi.org/10.1038/s42003-019-0541-0
- Primary Citation of Related Structures:
6I50 - PubMed Abstract:
The tumour necrosis factor alpha (TNFα) superfamily of proteins are critical in numerous biological processes, such as in development and immunity. Eiger is the sole TNFα member described in arthropods such as in the important model organism Drosophila . To date there are no structural data on any Eiger protein. Here we present the structure of the TNF domain of Eiger from the fall armyworm Spodoptera frugiperda (SfEiger) to 1.7 Å from a serendipitously obtained crystal without prior knowledge of the protein sequence. Our structure confirms that canonical trimerization is conserved from ancestral TNFs and points towards a mode of receptor engagement. Furthermore, we observe numerous surface histidines on SfEiger, potentially acting as pH switches following internalization into endosomes. Our data contributes to the genome annotation of S. frugiperda , a voracious agricultural pest, and can serve as a basis for future structure-function investigations of the TNF system in related arthropods such as Drosophila .
Organizational Affiliation:
1Division of Structural Biology, Wellcome Centre for Human Genetics, University of Oxford, 10 Roosevelt Drive, Oxford, OX3 7BN UK.