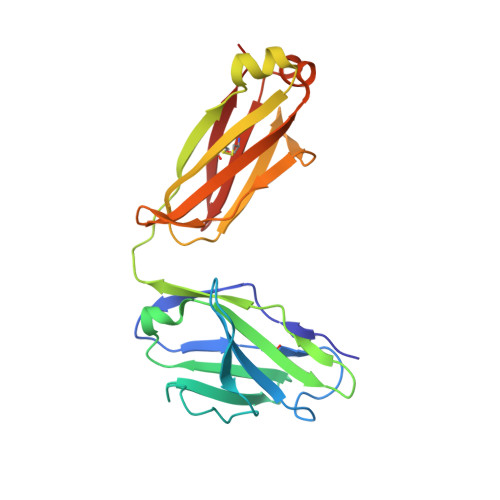
Structural basis for recognition of the malaria vaccine candidate Pfs48/45 by a transmission blocking antibody.
Lennartz, F., Brod, F., Dabbs, R., Miura, K., Mekhaiel, D., Marini, A., Jore, M.M., Sogaard, M.M., Jorgensen, T., de Jongh, W.A., Sauerwein, R.W., Long, C.A., Biswas, S., Higgins, M.K.(2018) Nat Commun 9: 3822-3822
- PubMed: 30237518
- DOI: https://doi.org/10.1038/s41467-018-06340-9
- Primary Citation of Related Structures:
6H5N - PubMed Abstract:
The quest to develop an effective malaria vaccine remains a major priority in the fight against global infectious disease. An approach with great potential is a transmission-blocking vaccine which induces antibodies that prevent establishment of a productive infection in mosquitos that feed on infected humans, thereby stopping the transmission cycle. One of the most promising targets for such a vaccine is the gamete surface protein, Pfs48/45. Here we establish a system for production of full-length Pfs48/45 and use this to raise a panel of monoclonal antibodies. We map the binding regions of these antibodies on Pfs48/45 and correlate the location of their epitopes with their transmission-blocking activity. Finally, we present the structure of the C-terminal domain of Pfs48/45 bound to the most potent transmission-blocking antibody, and provide key molecular information for future structure-guided immunogen design.
Organizational Affiliation:
Department of Biochemistry, University of Oxford, South Parks Road, Oxford, OX1 3QU, UK.