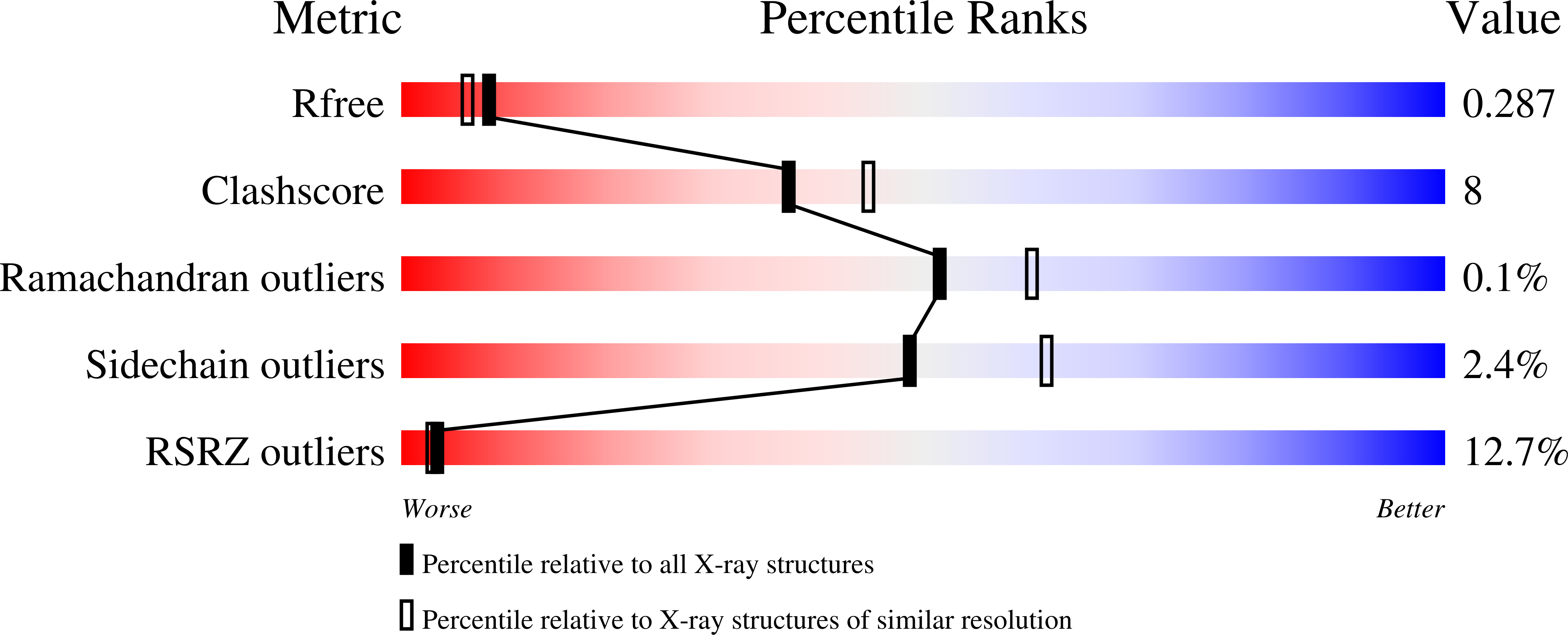
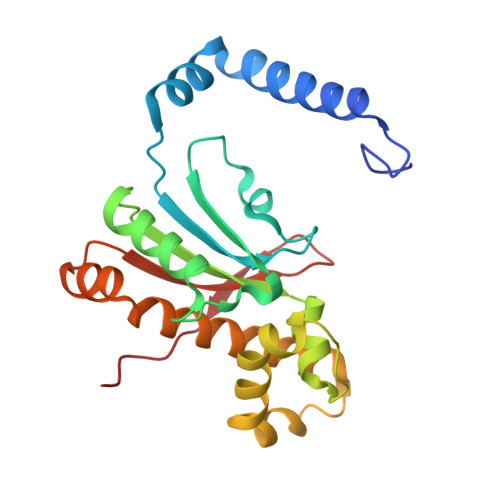
Crystal structure of carbonic anhydrase CaNce103p from the pathogenic yeast Candida albicans.
Dostal, J., Brynda, J., Blaha, J., Machacek, S., Heidingsfeld, O., Pichova, I.(2018) BMC Struct Biol 18: 14-14
- PubMed: 30367660
- DOI: https://doi.org/10.1186/s12900-018-0093-4
- Primary Citation of Related Structures:
6GWU - PubMed Abstract:
The pathogenic yeast Candida albicans can proliferate in environments with different carbon dioxide concentrations thanks to the carbonic anhydrase CaNce103p, which accelerates spontaneous conversion of carbon dioxide to bicarbonate and vice versa. Without functional CaNce103p, C. albicans cannot survive in atmospheric air. CaNce103p falls into the β-carbonic anhydrase class, along with its ortholog ScNce103p from Saccharomyces cerevisiae. The crystal structure of CaNce103p is of interest because this enzyme is a potential target for surface disinfectants.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Flemingovo náměstí 2, 166 10, Prague, Czech Republic.