A Molecular Hybridization Approach for the Design of Potent, Highly Selective, and Brain-Penetrant N-Myristoyltransferase Inhibitors.
Harrison, J.R., Brand, S., Smith, V., Robinson, D.A., Thompson, S., Smith, A., Davies, K., Mok, N., Torrie, L.S., Collie, I., Hallyburton, I., Norval, S., Simeons, F.R.C., Stojanovski, L., Frearson, J.A., Brenk, R., Wyatt, P.G., Gilbert, I.H., Read, K.D.(2018) J Med Chem 61: 8374-8389
- PubMed: 30207721
- DOI: https://doi.org/10.1021/acs.jmedchem.8b00884
- Primary Citation of Related Structures:
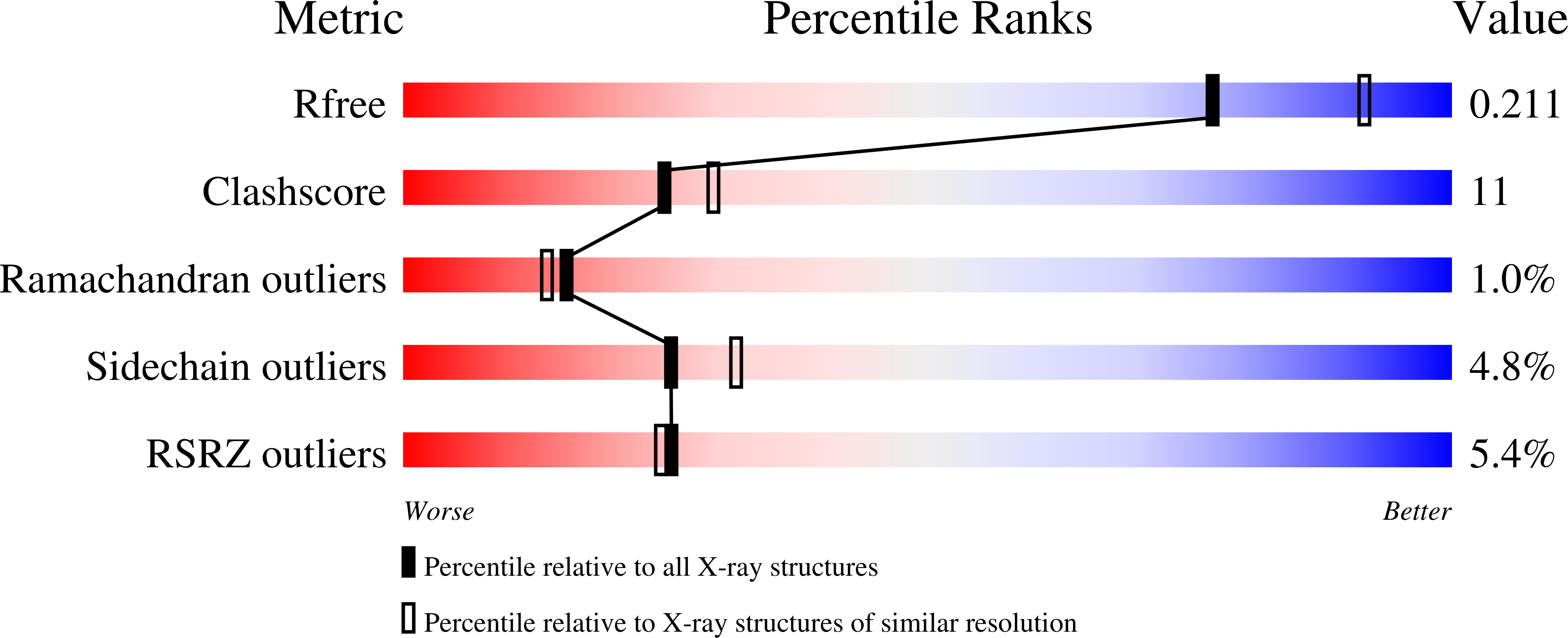
6GNH, 6GNS, 6GNT, 6GNU, 6GNV - PubMed Abstract:
Crystallography has guided the hybridization of two series of Trypanosoma brucei N-myristoyltransferase (NMT) inhibitors, leading to a novel highly selective series. The effect of combining the selectivity enhancing elements from two pharmacophores is shown to be additive and has led to compounds that have greater than 1000-fold selectivity for TbNMT vs HsNMT. Further optimization of the hybrid series has identified compounds with significant trypanocidal activity capable of crossing the blood-brain barrier. By using CF-1 mdr1a deficient mice, we were able to demonstrate full cures in vivo in a mouse model of stage 2 African sleeping sickness. This and previous work provides very strong validation for NMT as a drug target for human African trypanosomiasis in both the peripheral and central nervous system stages of disease.
Organizational Affiliation:
Drug Discovery Unit, Wellcome Centre for Anti-Infectives Research, Division of Biological Chemistry and Drug Discovery, School of Life Sciences , University of Dundee , Dundee , DD1 5EH , United Kingdom.