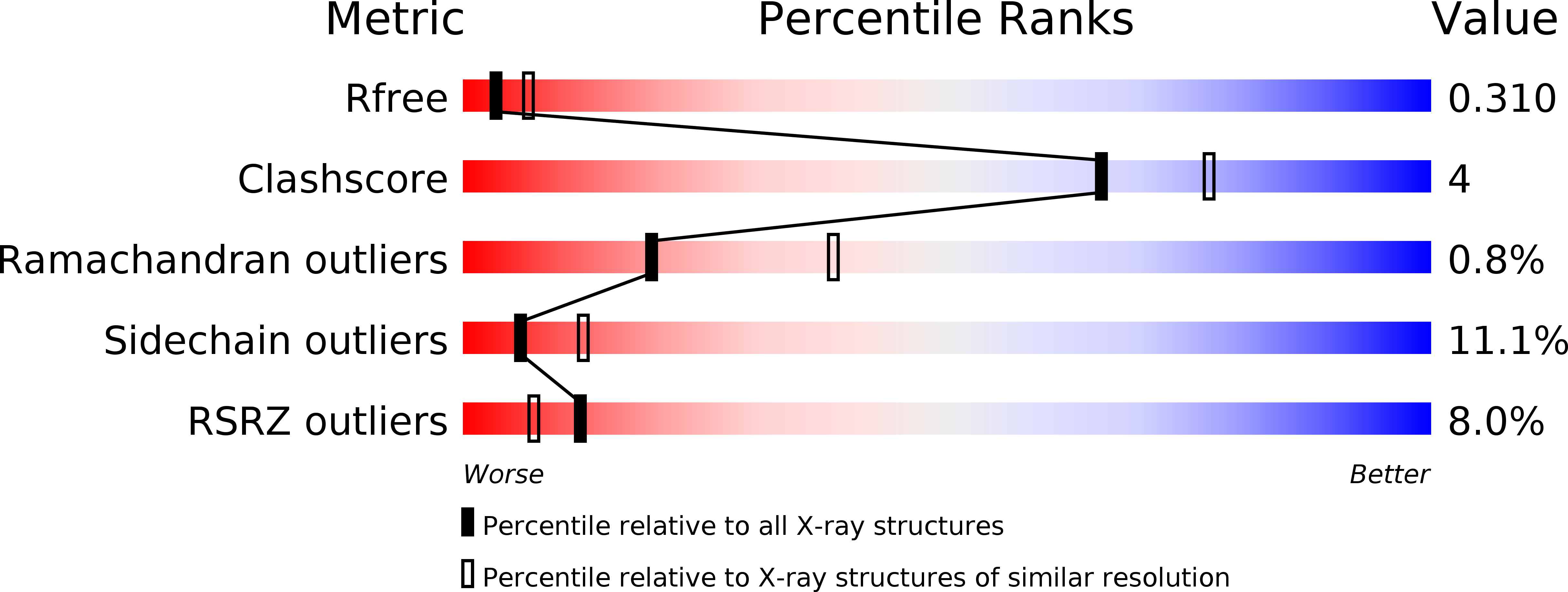
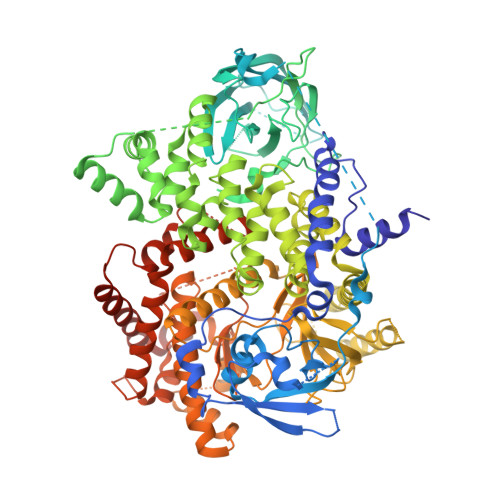
Discovery and Optimisation of a Novel Series of 3-Cinnoline Carboxamides as Orally Bioavailable, Highly Potent and Selective Ataxia Telangiectasia Mutated (ATM) inhibitors
Barlaam, B., Cadogan, E., Cambell, A., Colclough, N., Dishington, A., Durant, S., Goldberg, K., Hassall, L.A., Hughes, G.D., MacFaul, P.A., McGuire, T.M., Pass, M., Patel, A., Pearson, S., Petersen, J., Pike, K.G., Robb, G., Stratton, N., Xin, N., Zhai, B.To be published.