Tyrosine metabolism: identification of a key residue in the acquisition of prephenate aminotransferase activity by 1 beta aspartate aminotransferase.
Giustini, C., Graindorge, M., Cobessi, D., Crouzy, S., Robin, A., Curien, G., Matringe, M.(2019) FEBS J 286: 2118-2134
- PubMed: 30771275
- DOI: https://doi.org/10.1111/febs.14789
- Primary Citation of Related Structures:
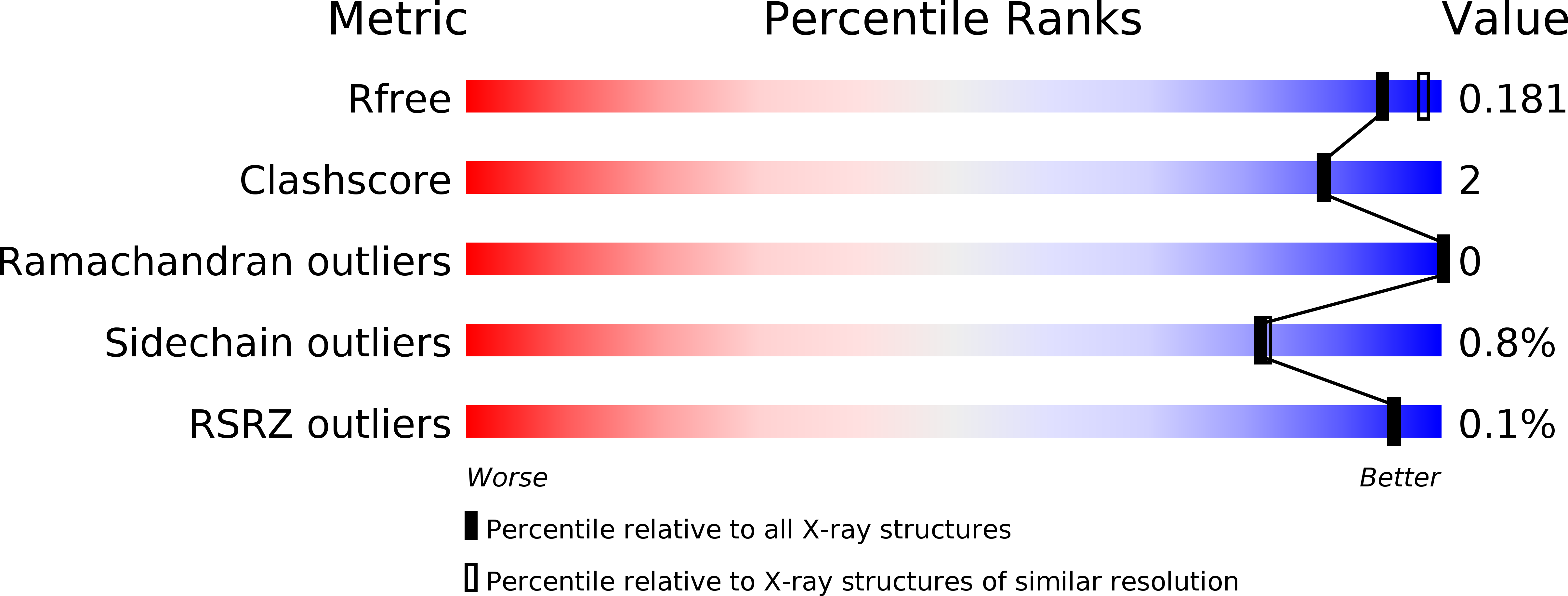
6F35, 6F5V, 6F77 - PubMed Abstract:
Alternative routes for the post-chorismate branch of the biosynthetic pathway leading to tyrosine exist, the 4-hydroxyphenylpyruvate or the arogenate route. The arogenate route involves the transamination of prephenate into arogenate. In a previous study, we found that, depending on the microorganisms possessing the arogenate route, three different aminotransferases evolved to perform prephenate transamination, that is, 1β aspartate aminotransferase (1β AAT), N-succinyl-l,l-diaminopimelate aminotransferase, and branched-chain aminotransferase. The present work aimed at identifying molecular determinant(s) of 1β AAT prephenate aminotransferase (PAT) activity. To that purpose, we conducted X-ray crystal structure analysis of two PAT competent 1β AAT from Arabidopsis thaliana and Rhizobium meliloti and one PAT incompetent 1β AAT from R. meliloti. This structural analysis supported by site-directed mutagenesis, modeling, and molecular dynamics simulations allowed us to identify a molecular determinant of PAT activity in the flexible N-terminal loop of 1β AAT. Our data reveal that a Lys/Arg/Gln residue in position 12 in the sequence (numbering according to Thermus thermophilus 1β AAT), present only in PAT competent enzymes, could interact with the 4-hydroxyl group of the prephenate substrate, and thus may have a central role in the acquisition of PAT activity by 1β AAT.
Organizational Affiliation:
INRA, CNRS, CEA, BIG-LPCV, University of Grenoble Alpes, France.