Crystal structures of a llama VHH antibody BCD090-M2 targeting human ErbB3 receptor.
Eliseev, I.E., Yudenko, A.N., Vysochinskaya, V.V., Svirina, A.A., Evstratyeva, A.V., Drozhzhachih, M.S., Krendeleva, E.A., Vladimirova, A.K., Nemankin, T.A., Ekimova, V.M., Ulitin, A.B., Lomovskaya, M.I., Yakovlev, P.A., Bukatin, A.S., Knyazev, N.A., Moiseenko, F.V., Chakchir, O.B.(2018) F1000Res 7: 57-57
- PubMed: 30430004
- DOI: https://doi.org/10.12688/f1000research.13612.2
- Primary Citation of Related Structures:
6EZW, 6F0D - PubMed Abstract:
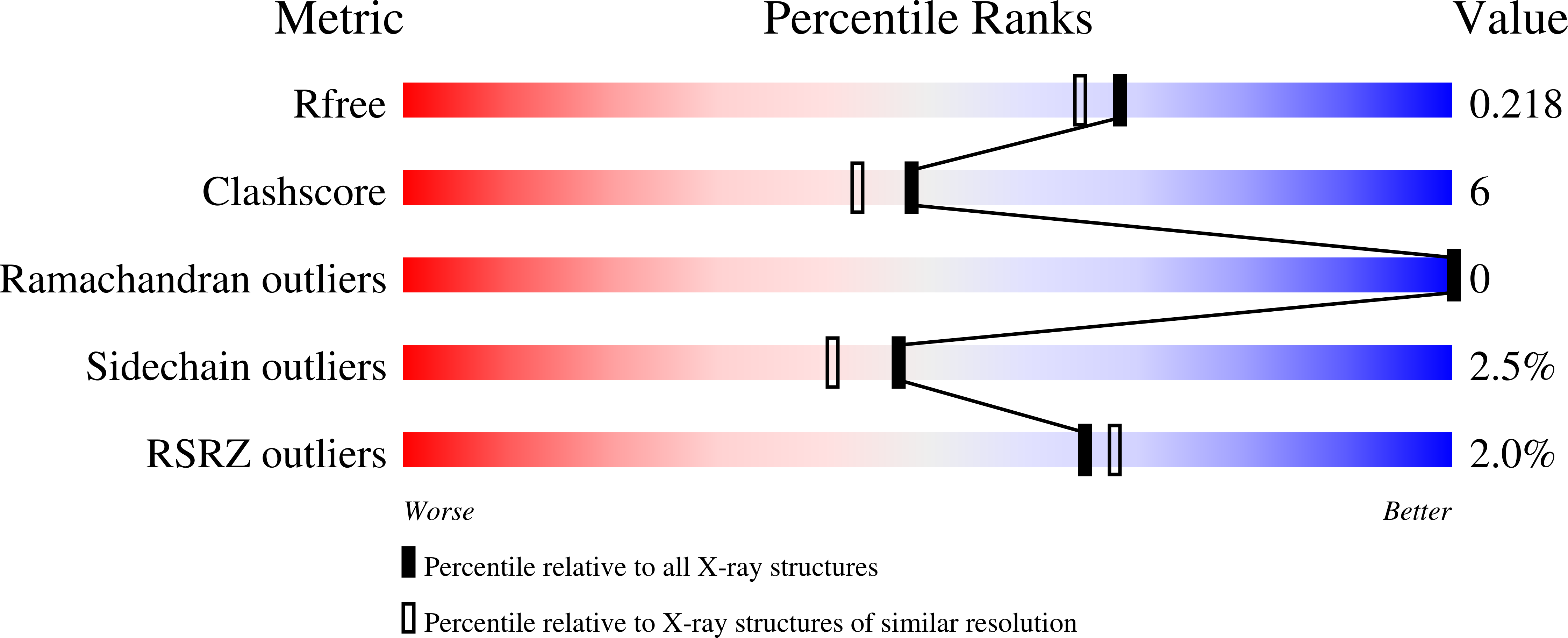
Background : The ability of ErbB3 receptor to functionally complement ErbB1-2 and induce tumor resistance to their inhibitors makes it a unique target in cancer therapy by monoclonal antibodies. Here we report the expression, purification and structural analysis of a new anti-ErbB3 single-chain antibody. Methods : The VHH fragment of the antibody was expressed in E. coli SHuffle cells as a SUMO fusion, cleaved by TEV protease and purified to homogeneity. Binding to the extracellular domain of ErbB3 was studied by surface plasmon resonance. For structural studies, the antibody was crystallized by hanging-drop vapor diffusion in two different forms. Results : We developed a robust and efficient system for recombinant expression of single-domain antibodies. The purified antibody was functional and bound ErbB3 with K D =15±1 nM. The crystal structures of the VHH antibody in space groups C2 and P1 were solved by molecular replacement at 1.6 and 1.9 Å resolution. The high-quality electron density maps allowed us to build precise atomic models of the antibody and the putative paratope. Surprisingly, the CDR H2 existed in multiple distant conformations in different crystal forms, while the more complex CDR H3 had a low structural variability. The structures were deposited under PDB entry codes 6EZW and 6F0D. Conclusions : Our results may facilitate further mechanistic studies of ErbB3 inhibition by single-chain antibodies. Besides, the solved structures will contribute to datasets required to develop new computational methods for antibody modeling and design.
Organizational Affiliation:
St. Petersburg National Research Academic University RAS, St. Petersburg, 194021, Russian Federation.