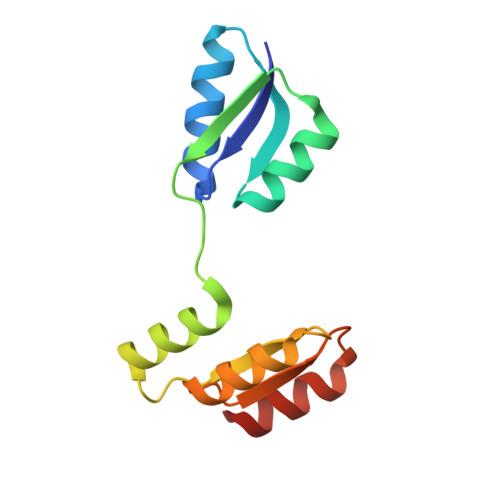
Structure of the archaeal chemotaxis protein CheY in a domain-swapped dimeric conformation.
Paithankar, K.S., Enderle, M., Wirthensohn, D.C., Miller, A., Schlesner, M., Pfeiffer, F., Rittner, A., Grininger, M., Oesterhelt, D.(2019) Acta Crystallogr F Struct Biol Commun 75: 576-585
- PubMed: 31475924
- DOI: https://doi.org/10.1107/S2053230X19010896
- Primary Citation of Related Structures:
6ER7, 6EXR - PubMed Abstract:
Archaea are motile by the rotation of the archaellum. The archaellum switches between clockwise and counterclockwise rotation, and movement along a chemical gradient is possible by modulation of the switching frequency. This modulation involves the response regulator CheY and the archaellum adaptor protein CheF. In this study, two new crystal forms and protein structures of CheY are reported. In both crystal forms, CheY is arranged in a domain-swapped conformation. CheF, the protein bridging the chemotaxis signal transduction system and the motility apparatus, was recombinantly expressed, purified and subjected to X-ray data collection.
Organizational Affiliation:
Institute of Organic Chemistry and Chemical Biology, Buchmann Institute for Molecular Life Sciences, Goethe University Frankfurt, Max-von-Laue-Strasse 15, 60438 Frankfurt am Main, Germany.