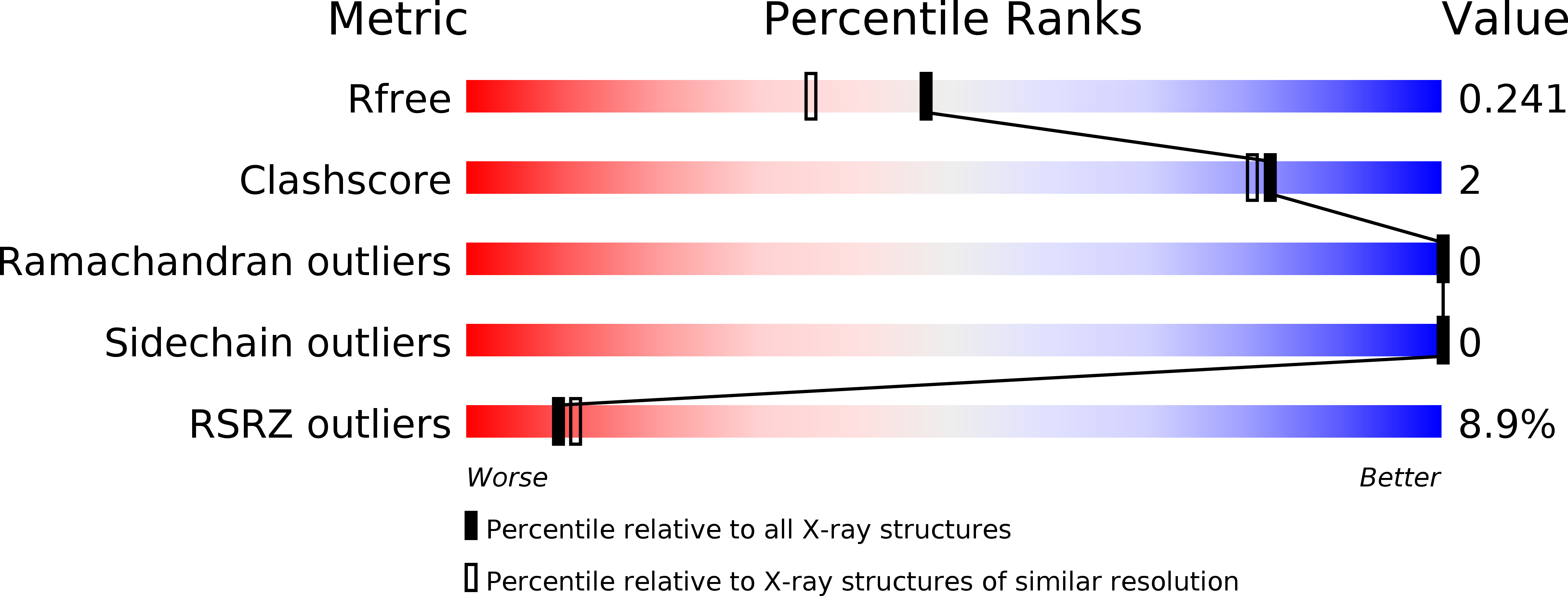
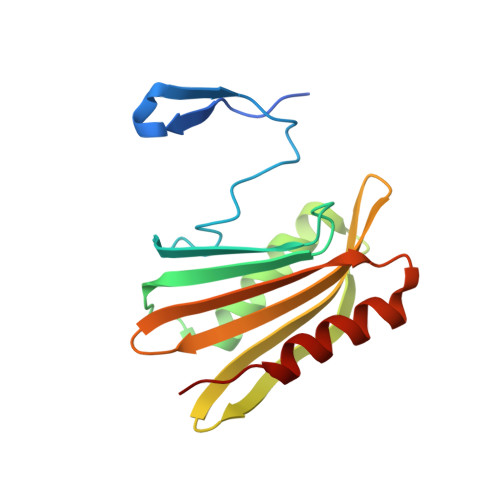
The structure of DcrB, a lipoprotein from Salmonella enterica, reveals flexibility in the N-terminal segment of the Mog1p/PsbP-like fold.
Rasmussen, D.M., Soens, R.W., Davie, T.J., Vaneerd, C.K., Bhattacharyya, B., May, J.F.(2018) J Struct Biol 204: 513-518
- PubMed: 30339832
- DOI: https://doi.org/10.1016/j.jsb.2018.10.005
- Primary Citation of Related Structures:
6E8A - PubMed Abstract:
DcrB is an 18 kDa lipoprotein that contains a single domain of unknown function. DcrB is found within Enterobacteriaceae, a family of Gram-negative bacteria which includes pathogens that can cause food-borne illness and hospital-acquired infections. In Salmonella enterica serovar Typhimurium, DcrB is up-regulated by conditions that promote the production of known virulence factors. We determined the structure of a truncated form of DcrB from Salmonella to 1.92 Å resolution by X-ray crystallography. This truncated form, DcrBΔ37, contains the entire domain of unknown function but lacks the lipoprotein signal sequence (residues 1-20) as well as residues 21-37. The DcrBΔ37 monomer contains the Mog1p/PsbP-like fold, which is found in functionally diverse proteins in mammals, yeast, plants, and cyanobacteria. Interestingly, DcrBΔ37 crystallized as a domain-swapped homodimer in which the N-terminal β-hairpin extends from one protomer to interact with the core of the second protomer. This domain-swapping indicates that the N-terminal portion of the Mog1p/PsbP-like fold likely has conformational flexibility. Overall, our results provide the first example of an enterobacterial protein that contains the Mog1p/PsbP-like fold and expands knowledge of the structural and phylogenetic diversity of Mog1p/PsbP-like proteins.
Organizational Affiliation:
Department of Chemistry and Biochemistry, University of Wisconsin-La Crosse, 1725 State Street, La Crosse, WI 54601, United States.