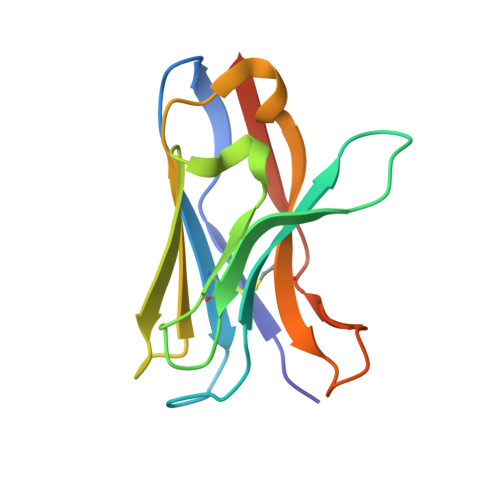
Structure of a VHH isolated from a naive phage display library.
White, B., Huh, I., Brooks, C.L.(2019) BMC Res Notes 12: 154-154
- PubMed: 30890176
- DOI: https://doi.org/10.1186/s13104-019-4197-0
- Primary Citation of Related Structures:
6DYX - PubMed Abstract:
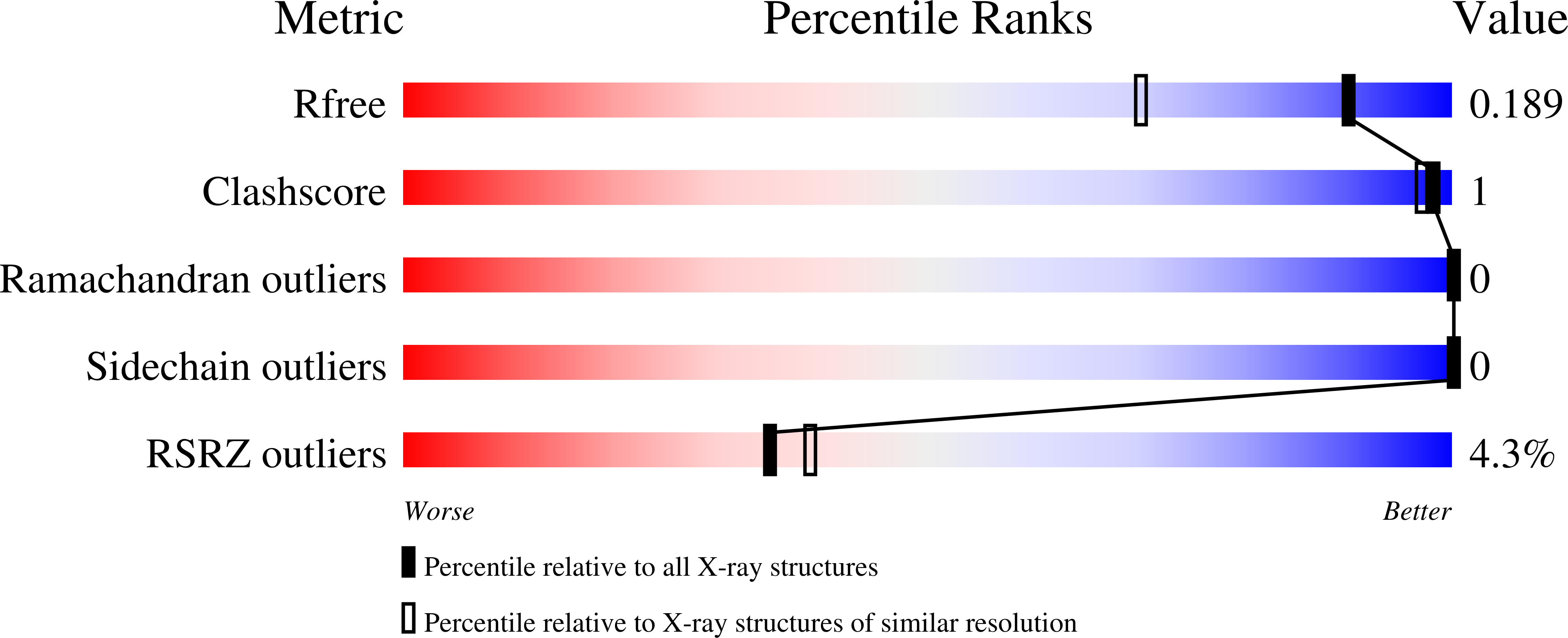
To determine the X-ray structure and biophysical properties of a Camelid V H H isolated from a naïve phage display library. Single domain antibodies (V H H) derived from the unique immune system of the Camelidae family have gained traction as useful tools for biotechnology as well as a source of potentially novel therapeutics. Here we report the structure and biophysical characterization of a V H H originally isolated from a naïve camelid phage display library. V H H R419 has a melting temperate of 66 °C and was found to be a monomer in solution. The protein crystallized in space group P6 5 22 and the structure was solved by molecular replacement to a resolution of 1.5 Å. The structure revealed a flat paratope with CDR loops that could be classified into existing canonical loop structures. A combination of high expression yield, stability and rapid crystallization might make R419 into a candidate scaffold for CDR grafting and homology modeling.
Organizational Affiliation:
Department of Chemistry, California State University Fresno, 2555 E San Ramon Ave, Fresno, CA, 93740, USA.