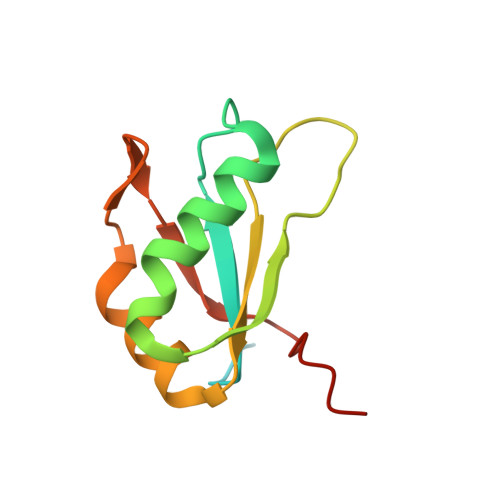
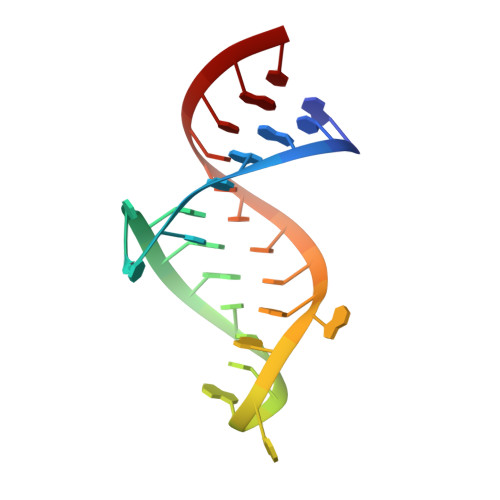
Structure of HIV TAR in complex with a Lab-Evolved RRM provides insight into duplex RNA recognition and synthesis of a constrained peptide that impairs transcription.
Belashov, I.A., Crawford, D.W., Cavender, C.E., Dai, P., Beardslee, P.C., Mathews, D.H., Pentelute, B.L., McNaughton, B.R., Wedekind, J.E.(2018) Nucleic Acids Res 46: 6401-6415
- PubMed: 29961805
- DOI: https://doi.org/10.1093/nar/gky529
- Primary Citation of Related Structures:
6CMN - PubMed Abstract:
Natural and lab-evolved proteins often recognize their RNA partners with exquisite affinity. Structural analysis of such complexes can offer valuable insight into sequence-selective recognition that can be exploited to alter biological function. Here, we describe the structure of a lab-evolved RNA recognition motif (RRM) bound to the HIV-1 trans-activation response (TAR) RNA element at 1.80 Å-resolution. The complex reveals a trio of arginines in an evolved β2-β3 loop penetrating deeply into the major groove to read conserved guanines while simultaneously forming cation-π and salt-bridge contacts. The observation that the evolved RRM engages TAR within a double-stranded stem is atypical compared to most RRMs. Mutagenesis, thermodynamic analysis and molecular dynamics validate the atypical binding mode and quantify molecular contributions that support the exceptionally tight binding of the TAR-protein complex (KD,App of 2.5 ± 0.1 nM). These findings led to the hypothesis that the β2-β3 loop can function as a standalone TAR-recognition module. Indeed, short constrained peptides comprising the β2-β3 loop still bind TAR (KD,App of 1.8 ± 0.5 μM) and significantly weaken TAR-dependent transcription. Our results provide a detailed understanding of TAR molecular recognition and reveal that a lab-evolved protein can be reduced to a minimal RNA-binding peptide.
Organizational Affiliation:
Department of Biochemistry & Biophysics, Center for RNA Biology, and Center for AIDS Research, University of Rochester School of Medicine & Dentistry, Rochester, NY 14642, USA.