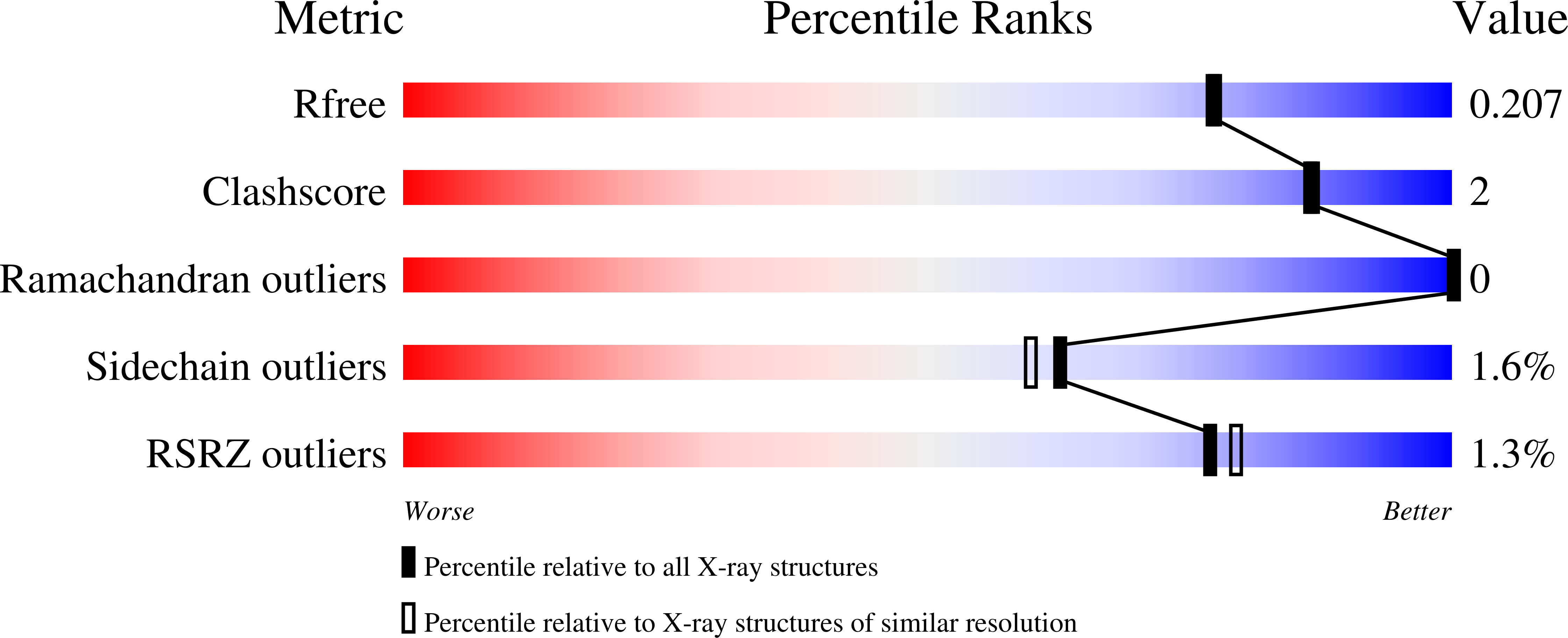
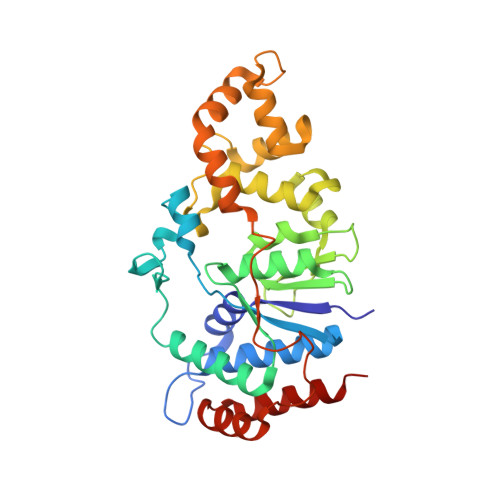
Crystal structure and mutational analysis of Mycobacterium smegmatis FenA highlight active site amino acids and three metal ions essential for flap endonuclease and 5' exonuclease activities.
Uson, M.L., Carl, A., Goldgur, Y., Shuman, S.(2018) Nucleic Acids Res 46: 4164-4175
- PubMed: 29635474
- DOI: https://doi.org/10.1093/nar/gky238
- Primary Citation of Related Structures:
6C33, 6C34, 6C35, 6C36 - PubMed Abstract:
Mycobacterium smegmatis FenA is a nucleic acid phosphodiesterase with flap endonuclease and 5' exonuclease activities. The 1.8 Å crystal structure of FenA reported here highlights as its closest homologs bacterial FEN-family enzymes ExoIX, the Pol1 exonuclease domain and phage T5 Fen. Mycobacterial FenA assimilates three active site manganese ions (M1, M2, M3) that are coordinated, directly and via waters, to a constellation of eight carboxylate side chains. We find via mutagenesis that the carboxylate contacts to all three manganese ions are essential for FenA's activities. Structures of nuclease-dead FenA mutants D125N, D148N and D208N reveal how they fail to bind one of the three active site Mn2+ ions, in a distinctive fashion for each Asn change. The structure of FenA D208N with a phosphate anion engaged by M1 and M2 in a state mimetic of a product complex suggests a mechanism for metal-catalyzed phosphodiester hydrolysis similar to that proposed for human Exo1. A distinctive feature of FenA is that it does not have the helical arch module found in many other FEN/FEN-like enzymes. Instead, this segment of FenA adopts a unique structure comprising a short 310 helix and surface β-loop that coordinates a fourth manganese ion (M4).
Organizational Affiliation:
Molecular Biology Program, Sloan-Kettering Institute, New York, NY 10065, USA.