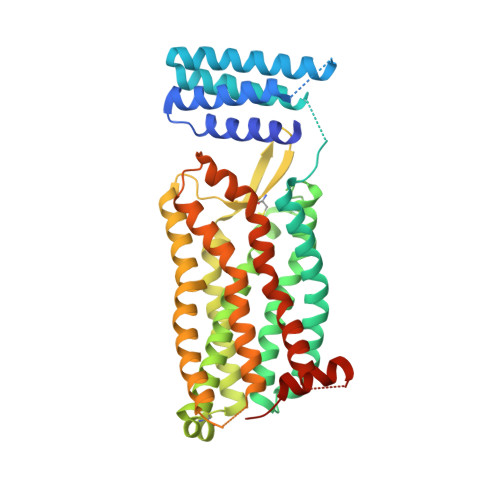
Orthosteric and allosteric action of the C5a receptor antagonists.
Liu, H., Kim, H.R., Deepak, R.N.V.K., Wang, L., Chung, K.Y., Fan, H., Wei, Z., Zhang, C.(2018) Nat Struct Mol Biol 25: 472-481
- PubMed: 29867214
- DOI: https://doi.org/10.1038/s41594-018-0067-z
- Primary Citation of Related Structures:
6C1Q, 6C1R - PubMed Abstract:
The C5a receptor (C5aR) is a G-protein-coupled receptor (GPCR) that can induce strong inflammatory response to the anaphylatoxin C5a. Targeting C5aR has emerged as a novel anti-inflammatory therapeutic method. However, developing potent C5aR antagonists as drugs has proven difficult. Here, we report two crystal structures of human C5aR in ternary complexes with the peptide antagonist PMX53 and a non-peptide antagonist, either avacopan or NDT9513727. The structures, together with other biophysical, computational docking and cell-based signaling data, reveal the orthosteric action of PMX53 and its effect of stabilizing the C5aR structure, as well as the allosteric action of chemically diverse non-peptide C5aR antagonists with different binding poses. Structural comparison analysis suggests the presence of similar allosteric sites in other GPCRs. We also discuss critical structural features of C5aR in activation, including a novel conformation of helix 8. On the basis of our results, we suggest novel strategies for developing C5aR-targeting drugs.
Organizational Affiliation:
Department of Pharmacology and Chemical Biology, School of Medicine, University of Pittsburgh, Pittsburgh, PA, USA.