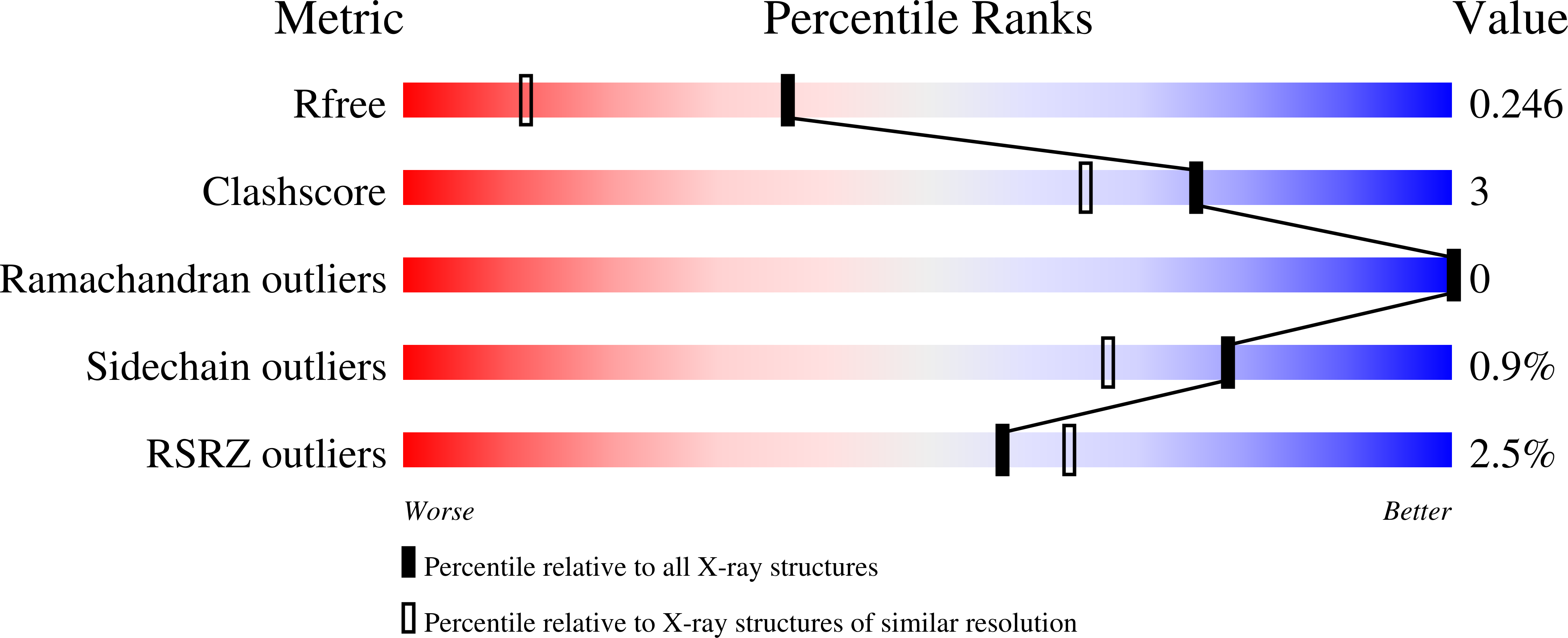
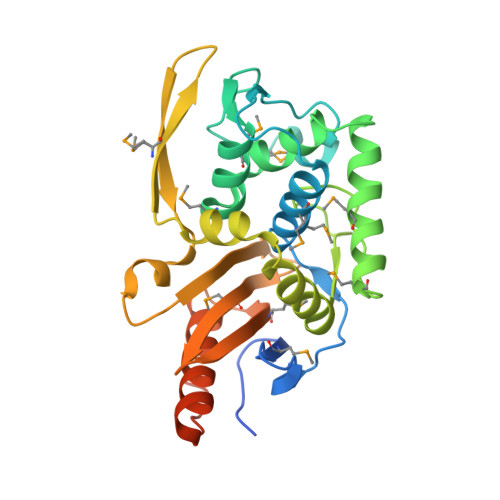
1.75 Angstrom Resolution Crystal Structure of D-alanyl-D-alanine Endopeptidase from Enterobacter cloacae in Complex with Covalently Bound Boronic Acid.
Minasov, G., Wawrzak, Z., Shuvalova, L., Kiryukhina, O., Dubrovska, I., Grimshaw, S., Kwon, K., Anderson, W.F., Satchell, K.J.F., Joachimiak, A.To be published.