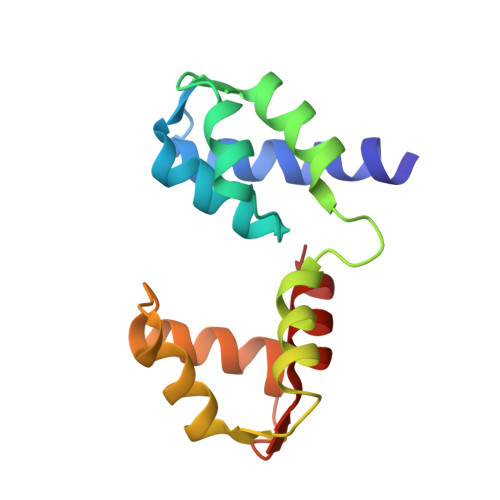
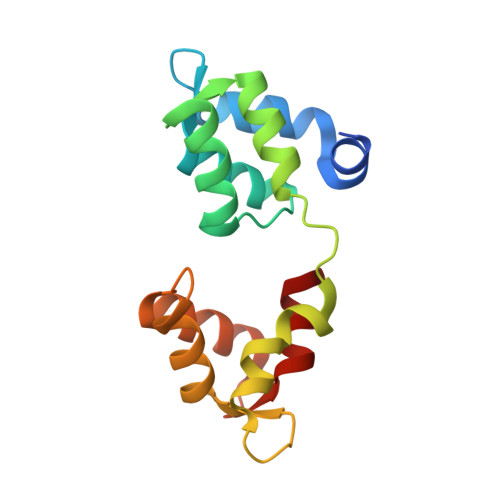
Structural role of essential light chains in the apicomplexan glideosome.
Pazicky, S., Dhamotharan, K., Kaszuba, K., Mertens, H.D.T., Gilberger, T., Svergun, D., Kosinski, J., Weininger, U., Low, C.(2020) Commun Biol 3: 568-568
- PubMed: 33051581
- DOI: https://doi.org/10.1038/s42003-020-01283-8
- Primary Citation of Related Structures:
6TJ3, 6TJ4, 6TJ5, 6TJ6, 6TJ7, 6ZN3 - PubMed Abstract:
Gliding, a type of motility based on an actin-myosin motor, is specific to apicomplexan parasites. Myosin A binds two light chains which further interact with glideosome associated proteins and assemble into the glideosome. The role of individual glideosome proteins is unclear due to the lack of structures of larger glideosome assemblies. Here, we investigate the role of essential light chains (ELCs) in Toxoplasma gondii and Plasmodium falciparum and present their crystal structures as part of trimeric sub-complexes. We show that although ELCs bind a conserved MyoA sequence, P. falciparum ELC adopts a distinct structure in the free and MyoA-bound state. We suggest that ELCs enhance MyoA performance by inducing secondary structure in MyoA and thus stiffen its lever arm. Structural and biophysical analysis reveals that calcium binding has no influence on the structure of ELCs. Our work represents a further step towards understanding the mechanism of gliding in Apicomplexa.
Organizational Affiliation:
Centre for Structural Systems Biology (CSSB), Notkestrasse 85, D-22607, Hamburg, Germany.