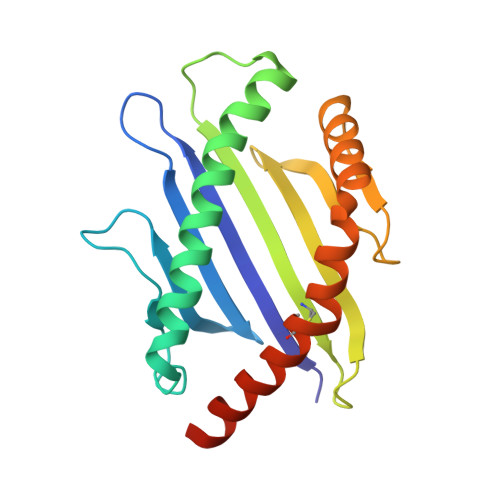
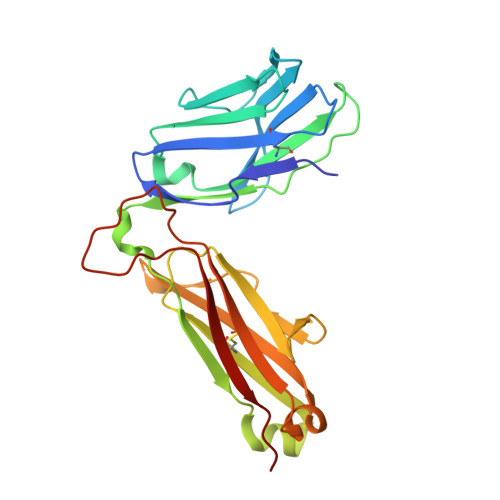
Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Li, X., Mizsei, R., Tan, K., Mallis, R.J., Duke-Cohan, J.S., Akitsu, A., Tetteh, P.W., Dubey, A., Hwang, W., Wagner, G., Lang, M.J., Arthanari, H., Wang, J.H., Reinherz, E.L.(2021) Science 371: 181-185
- PubMed: 33335016
- DOI: https://doi.org/10.1126/science.abe0918
- Primary Citation of Related Structures:
6WL2, 6WL3, 6WL4, 7JI2 - PubMed Abstract:
Self-discrimination, a critical but ill-defined molecular process programmed during thymocyte development, requires myriad pre-T cell receptors (preTCRs) and αβTCRs. Using x-ray crystallography, we show how a preTCR applies the concave β-sheet surface of its single variable domain (Vβ) to "horizontally" grab the protruding MHC α2-helix. By contrast, αβTCRs purpose all six complementarity-determining region (CDR) loops of their paired VαVβ module to recognize peptides bound to major histocompatibility complex molecules (pMHCs) in "vertical" head-to-head binding. The preTCR topological fit ensures that CDR3β reaches the peptide's featured C-terminal segment for pMHC sampling, establishing the subsequent αβTCR canonical docking mode. "Horizontal" docking precludes germline CDR1β- and CDR2β-MHC binding to broaden β-chain repertoire diversification before αβTCR-mediated selection refinement. Thus, one subunit successively attunes the recognition logic of related multicomponent receptors.
Organizational Affiliation:
Laboratory of Immunobiology, Dana-Farber Cancer Institute, Boston, MA, USA. jwang@crystal.harvard.edu ellis_reinherz@dfci.harvard.edu.