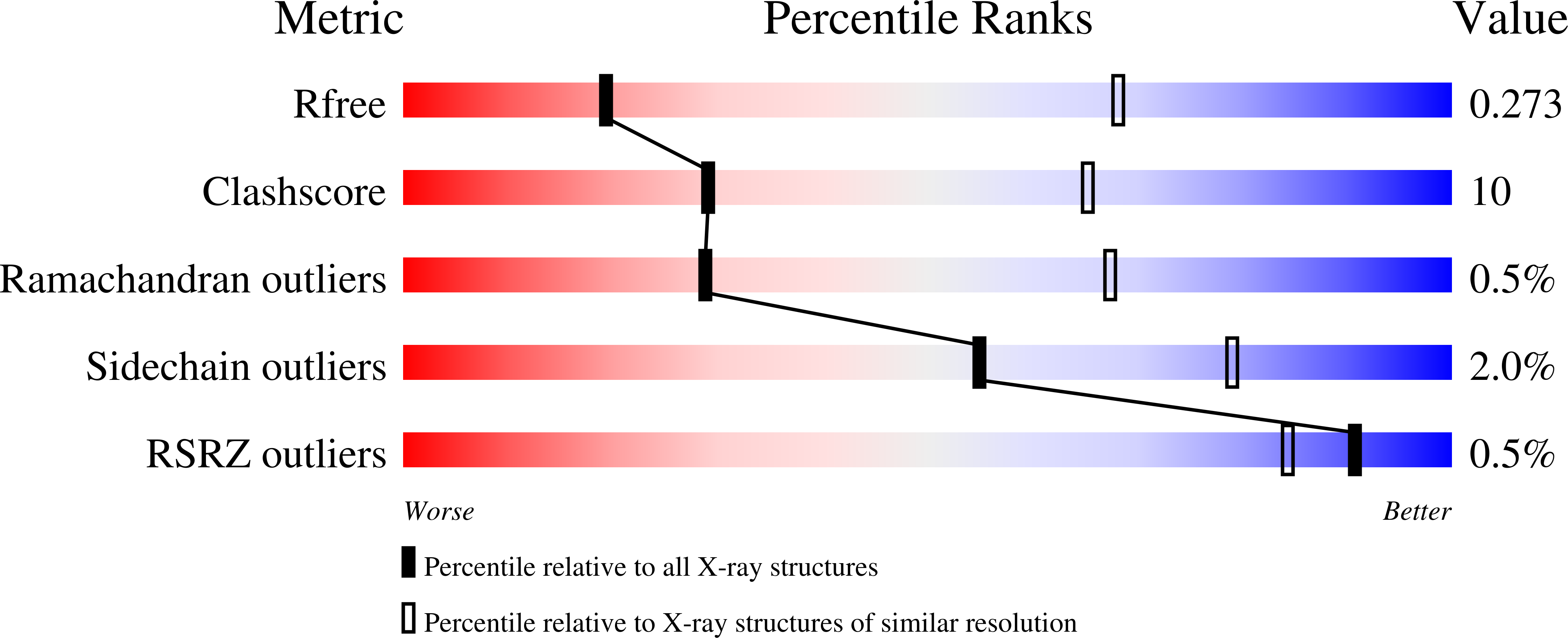
Structure and Mechanism of a Unique Diiron Center in Mammalian Stearoyl-CoA Desaturase.
Shen, J., Wu, G., Tsai, A.L., Zhou, M.(2020) J Mol Biol 432: 5152-5161
- PubMed: 32470559
- DOI: https://doi.org/10.1016/j.jmb.2020.05.017
- Primary Citation of Related Structures:
6WF2 - PubMed Abstract:
Stearoyl-CoA desaturase 1 (SCD1) is a membrane-embedded metalloenzyme that catalyzes the formation of a double bond on a saturated acyl-CoA. SCD1 has a diiron center and its proper function requires an electron transport chain composed of NADH (or NADPH), cytochrome b 5 reductase (b 5 R), and cytochrome b 5 (cyt b 5 ). Since SCD1 is a key regulator in fat metabolism and is required for survival of cancer cells, there is intense interest in targeting SCD1 for various metabolic diseases and cancers. Crystal structures of human and mouse SCD1 were reported recently; however, both proteins have two zinc ions instead of two iron ions in the catalytic center, and as a result, the enzymes are inactive. Here we report a general approach for incorporating iron into heterologously expressed proteins in HEK293 cells. We produced mouse SCD1 that contains a diiron center and visualized its diiron center by solving its crystal structure to 3.5 Å. We assembled the entire electron transport chain using the purified soluble domains of cyt b 5 and b 5 R, and the purified mouse SCD1, and we showed that three proteins coordinate to produce proper products. These results established an in vitro system that allows precise perturbations of the electron transport chain for the understanding of the catalytic mechanism in SCD1.
Organizational Affiliation:
Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA.