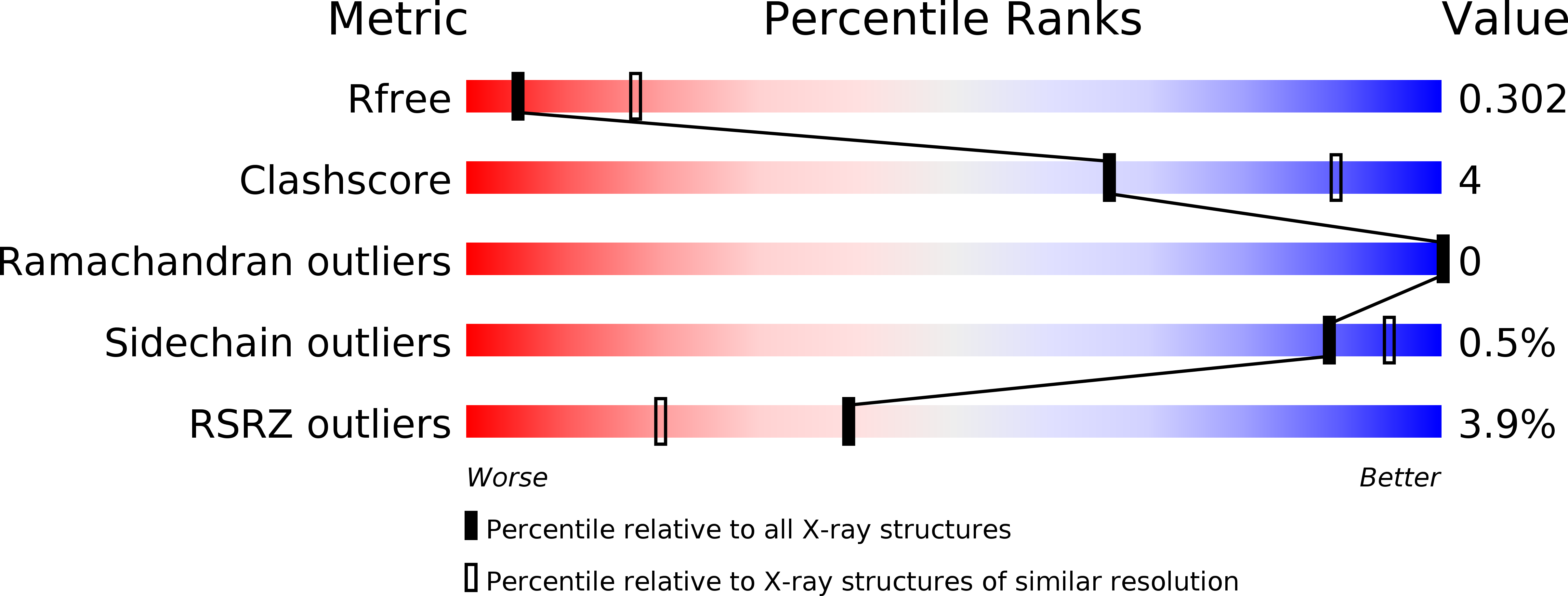
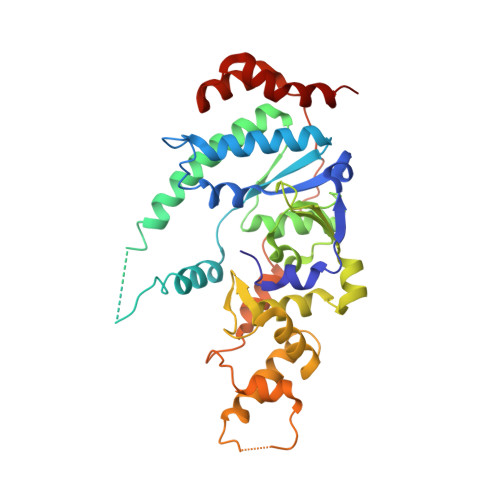
The crystal structure of human XPG, the xeroderma pigmentosum group G endonuclease, provides insight into nucleotide excision DNA repair.
Gonzalez-Corrochano, R., Ruiz, F.M., Taylor, N.M.I., Huecas, S., Drakulic, S., Spinola-Amilibia, M., Fernandez-Tornero, C.(2020) Nucleic Acids Res 48: 9943-9958
- PubMed: 32821917
- DOI: https://doi.org/10.1093/nar/gkaa688
- Primary Citation of Related Structures:
6TUR, 6TUS, 6TUW, 6TUX - PubMed Abstract:
Nucleotide excision repair (NER) is an essential pathway to remove bulky lesions affecting one strand of DNA. Defects in components of this repair system are at the ground of genetic diseases such as xeroderma pigmentosum (XP) and Cockayne syndrome (CS). The XP complementation group G (XPG) endonuclease cleaves the damaged DNA strand on the 3' side of the lesion coordinated with DNA re-synthesis. Here, we determined crystal structures of the XPG nuclease domain in the absence and presence of DNA. The overall fold exhibits similarities to other flap endonucleases but XPG harbors a dynamic helical arch that is uniquely oriented and defines a gateway. DNA binding through a helix-2-turn-helix motif, assisted by one flanking α-helix on each side, shows high plasticity, which is likely relevant for DNA scanning. A positively-charged canyon defined by the hydrophobic wedge and β-pin motifs provides an additional DNA-binding surface. Mutational analysis identifies helical arch residues that play critical roles in XPG function. A model for XPG participation in NER is proposed. Our structures and biochemical data represent a valuable tool to understand the atomic ground of XP and CS, and constitute a starting point for potential therapeutic applications.
Organizational Affiliation:
Centro de Investigaciones Biológicas Margarita Salas, CSIC, Ramiro de Maeztu 9, 28040 Madrid, Spain.